Input/Output#
In this section, we introduce how to input and output proxy data with cfr.
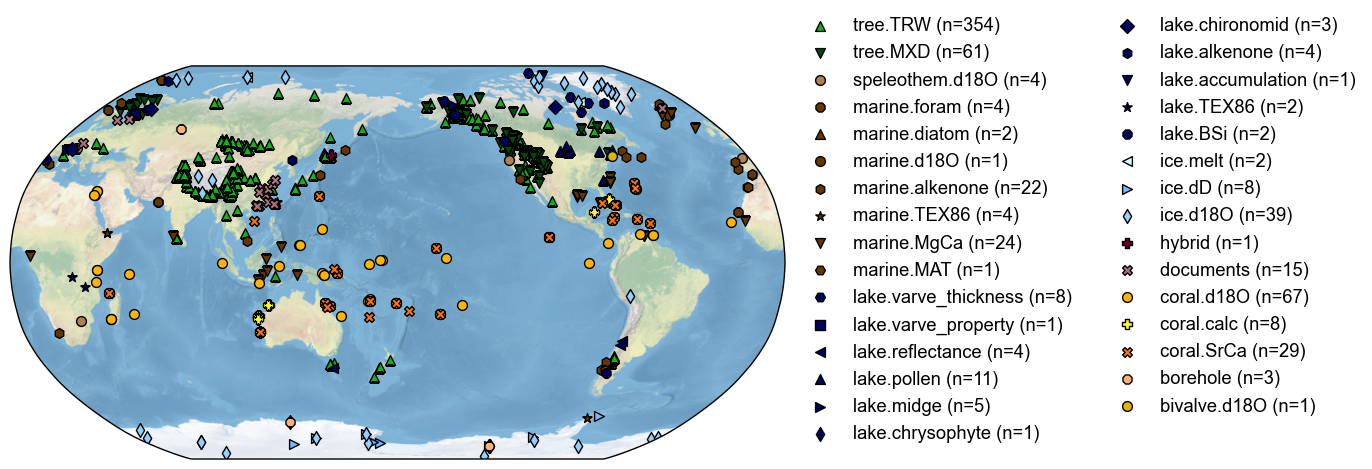
cfr provides a useful class called ProxyDatabase to conveniently store a collection of proxy records. Each record is stored in the form of a class called ProxyRecord. Here, we take the PAGES 2k global multiproxy database (PAGES2k Consortium, 2017) as an example to illustrate the basic usage of these two classes regarding data input/output.
Essentially, cfr supports below conversions:
pandas.DataFrame<=>cfr.ProxyDatabasea netCDF file <=>
cfr.ProxyDatabase(Experimental: this feature doesn’t support time axis before 1 CE)a netCDF file <=>
cfr.ProxyRecord(Experimental: this feature doesn’t support time axis before 1 CE)
In addition, cfr supports remote loading of hosted databases.
Required data to complete this tutorial:
PAGES2k Phase 2 database: pages2k_updated_Palmyra_dataset.pkl
[2]:
# import the packages we need for this tutorial
%load_ext autoreload
%autoreload 2
import cfr
print(cfr.__version__)
import xarray as xr
Remote loading databases#
cfr supports remote loading of hosted databases, currently including PAGES2kv2 and pseudoPAGES2k.
By calling the .fetch() method of ProxyDatabase without any arguments, a list of supported database names will be listed:
[3]:
pdb = cfr.ProxyDatabase().fetch()
>>> Choose one from the supported databases:
- PAGES2kv2
- pseudoPAGES2k/ppwn_SNRinf_rta
- pseudoPAGES2k/ppwn_SNR10_rta
- pseudoPAGES2k/ppwn_SNR2_rta
- pseudoPAGES2k/ppwn_SNR1_rta
- pseudoPAGES2k/ppwn_SNR0.5_rta
- pseudoPAGES2k/ppwn_SNR0.25_rta
- pseudoPAGES2k/ppwn_SNRinf_fta
- pseudoPAGES2k/ppwn_SNR10_fta
- pseudoPAGES2k/ppwn_SNR2_fta
- pseudoPAGES2k/ppwn_SNR1_fta
- pseudoPAGES2k/ppwn_SNR0.5_fta
- pseudoPAGES2k/ppwn_SNR0.25_fta
- pseudoPAGES2k/tpwn_SNR10_rta
- pseudoPAGES2k/tpwn_SNR2_rta
- pseudoPAGES2k/tpwn_SNR1_rta
- pseudoPAGES2k/tpwn_SNR0.5_rta
- pseudoPAGES2k/tpwn_SNR0.25_rta
- pseudoPAGES2k/tpwn_SNR10_fta
- pseudoPAGES2k/tpwn_SNR2_fta
- pseudoPAGES2k/tpwn_SNR1_fta
- pseudoPAGES2k/tpwn_SNR0.5_fta
- pseudoPAGES2k/tpwn_SNR0.25_fta
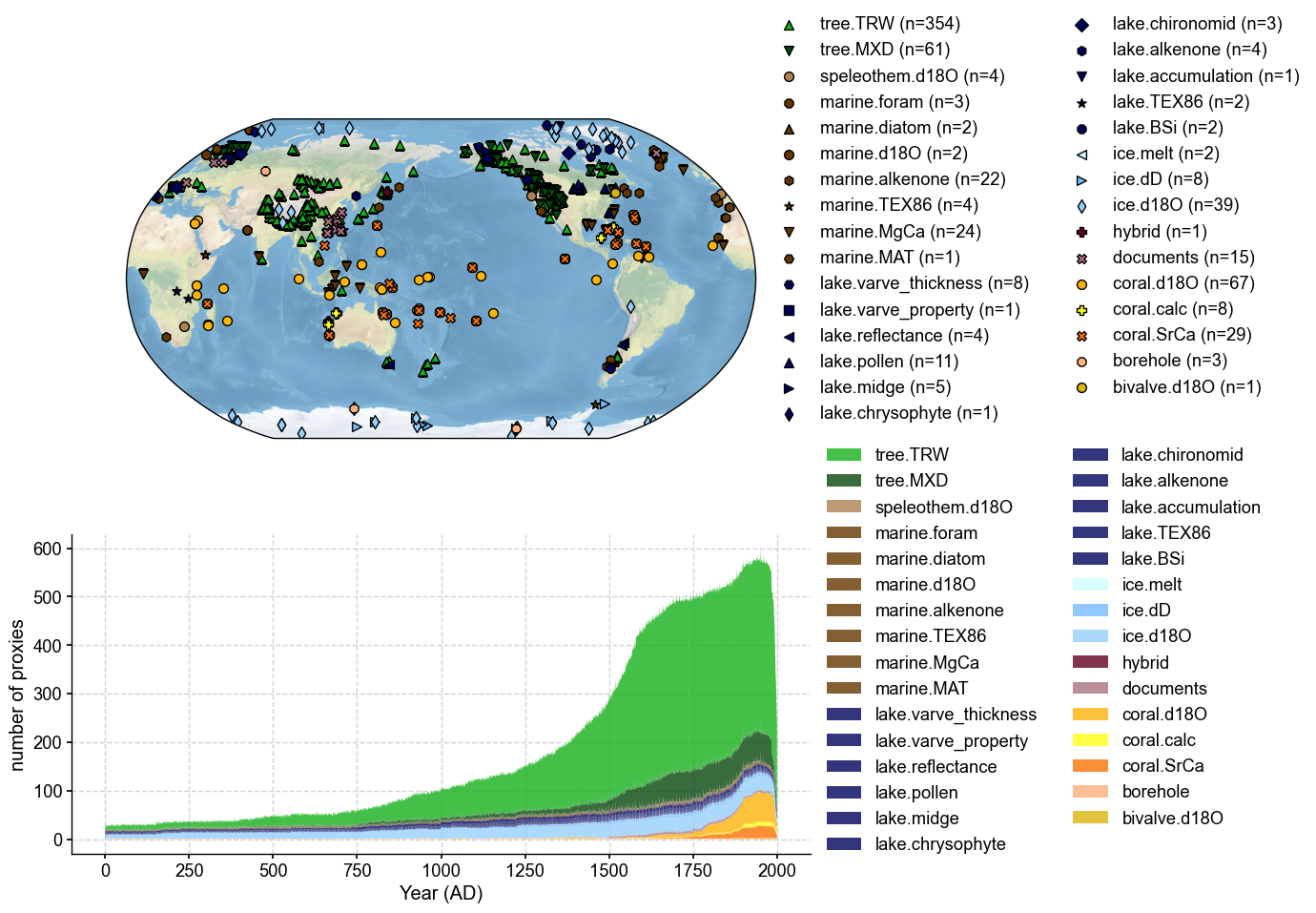
Remote loading PAGES2k#
[4]:
pdb = cfr.ProxyDatabase().fetch('PAGES2kv2')
fig, ax = pdb.plot(plot_count=True)
Remote loading pseudoPAGES2k#
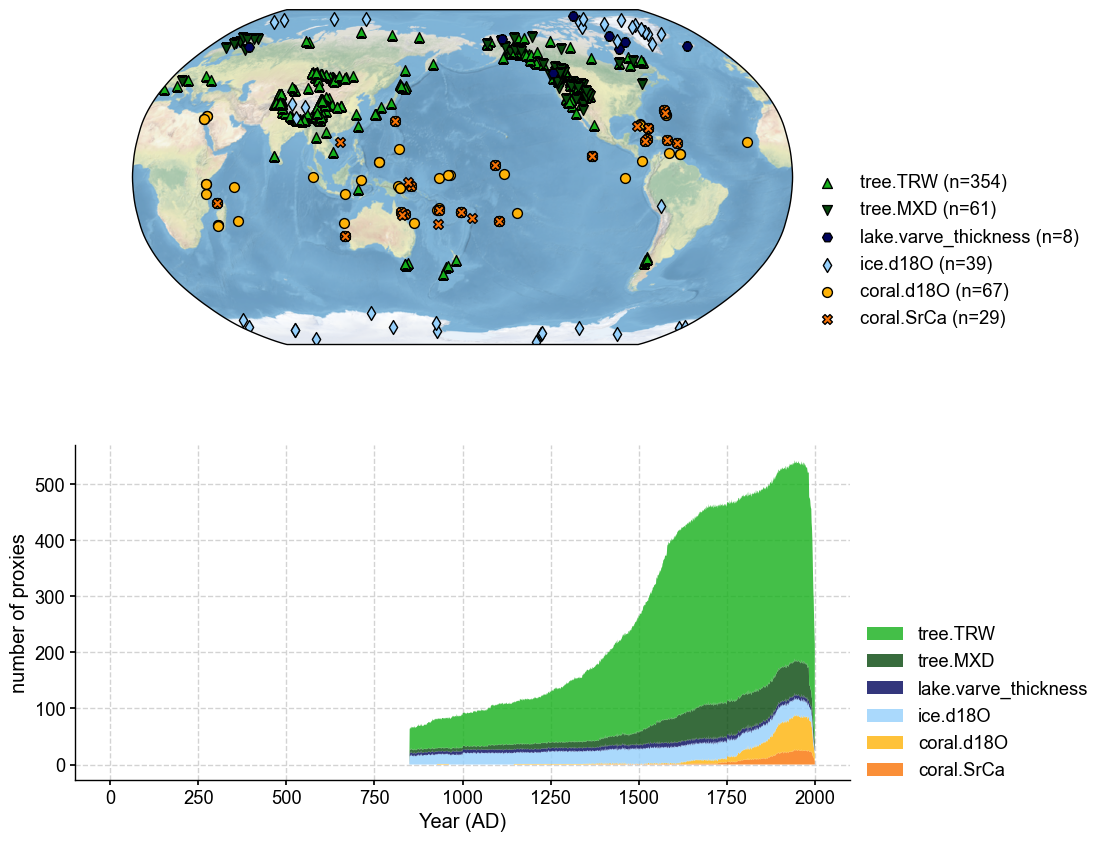
Note that there are different versions of pseudoPAEGS2k, such as “ppwn_SNRinf_rta” and “tpwn_SNR10_fta”. Those version names should be appended after “pseudoPAEGS2k/”. We show two examples below:
[5]:
pdb = cfr.ProxyDatabase().fetch('pseudoPAGES2k/ppwn_SNRinf_rta')
fig, ax = pdb.plot(plot_count=True)
[6]:
pdb = cfr.ProxyDatabase().fetch('pseudoPAGES2k/tpwn_SNR10_fta')
fig, ax = pdb.plot(plot_count=True)
cfr.ProxyDatabase => pandas.DataFrame#
Any cfr.ProxyDatabase can be converted to a pandas.DataFrame.
[12]:
df = pdb.to_df()
df
[12]:
| pid | lat | lon | elev | ptype | time | value | |
|---|---|---|---|---|---|---|---|
| 0 | NAm_153 | 52.7 | 241.7 | None | tree.TRW | [850.0, 851.0, 852.0, 853.0, 854.0, 855.0, 856... | [0.8828125, 1.03125, 1.0703125, 1.140625, 0.81... |
| 1 | Asi_245 | 23.0 | 114.0 | None | documents | [850.0, 851.0, 852.0, 853.0, 854.0, 855.0, 856... | [3.15625, 1.640625, 1.296875, 0.984375, -0.187... |
| 2 | NAm_165 | 37.9 | 252.3 | None | tree.MXD | [850.0, 851.0, 852.0, 853.0, 854.0, 855.0, 856... | [0.9765625, 1.03125, 1.0390625, 1.0390625, 0.9... |
| 3 | Asi_178 | 28.77 | 83.73 | None | tree.TRW | [850.0, 851.0, 852.0, 853.0, 854.0, 855.0, 856... | [1.3828125, 1.1328125, 1.296875, 1.109375, 0.8... |
| 4 | Asi_174 | 28.18 | 85.43 | None | tree.TRW | [850.0, 851.0, 852.0, 853.0, 854.0, 855.0, 856... | [1.3125, 1.1171875, 1.3125, 1.1484375, 0.77343... |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 687 | Asi_201 | 35.88 | 74.18 | None | tree.TRW | [850.0, 851.0, 852.0, 853.0, 854.0, 855.0, 856... | [1.3203125, 1.203125, 1.53125, 0.96875, 0.7109... |
| 688 | Asi_179 | 27.5 | 88.02 | None | tree.TRW | [850.0, 851.0, 852.0, 853.0, 854.0, 855.0, 856... | [1.3828125, 1.15625, 1.3203125, 1.140625, 0.82... |
| 689 | Arc_014 | 63.62 | 29.1 | None | lake.varve_thickness | [850.0, 851.0, 852.0, 853.0, 854.0, 855.0, 856... | [-1.046875, -0.7265625, -0.4765625, -1.2265625... |
| 690 | Ocn_071 | 16.2 | 298.51 | None | coral.d18O | [850.0, 851.0, 852.0, 853.0, 854.0, 855.0, 856... | [-3.9453125, -4.203125, -4.1640625, -3.84375, ... |
| 691 | Ocn_072 | 16.2 | 298.51 | None | coral.SrCa | [850.0, 851.0, 852.0, 853.0, 854.0, 855.0, 856... | [8.9375, 8.875, 8.8828125, 8.96875, 8.953125, ... |
692 rows × 7 columns
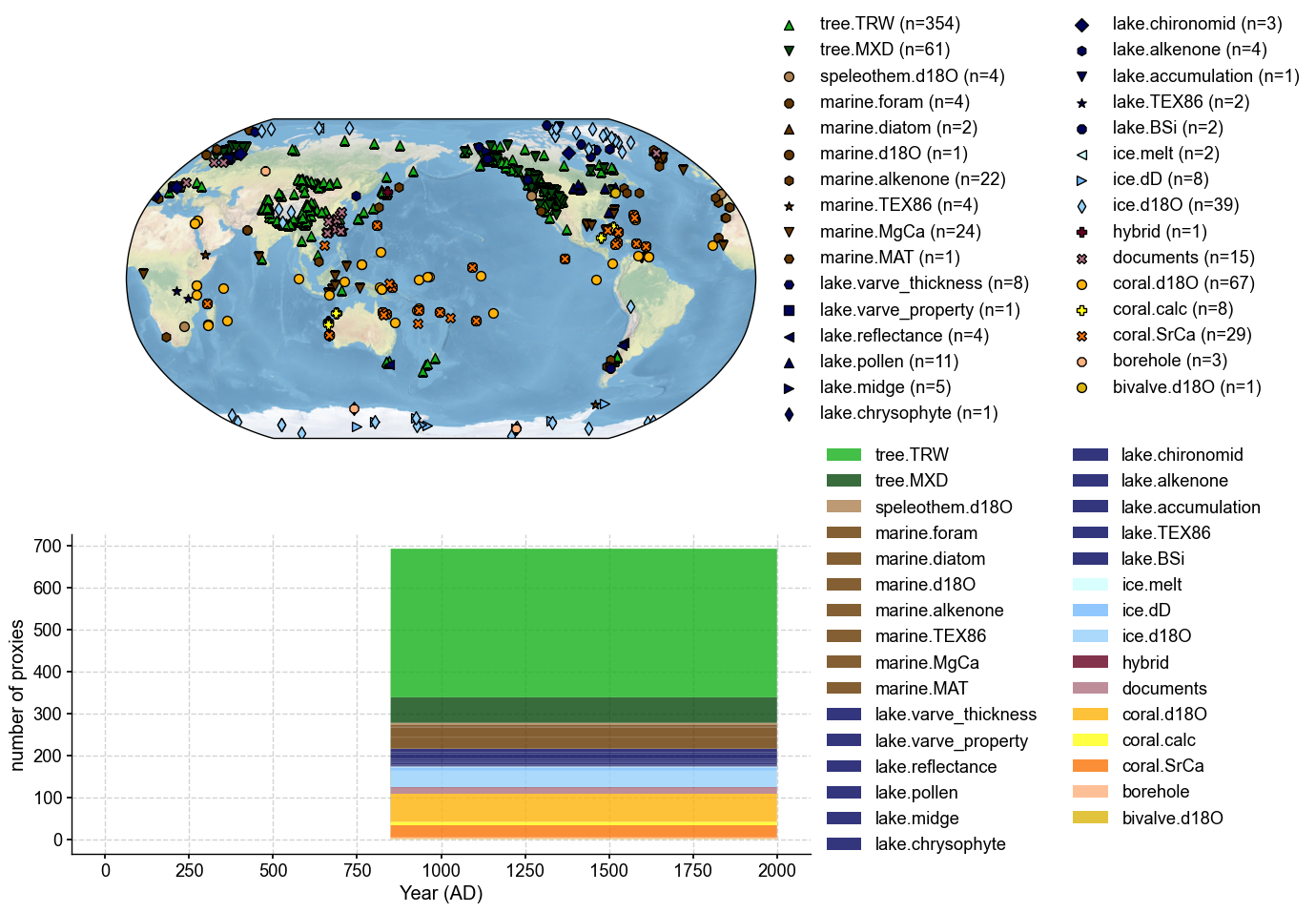
pandas.DataFrame => cfr.ProxyDatabase#
[11]:
pdb = cfr.ProxyDatabase().from_df(
df, pid_column='pid', lat_column='lat', lon_column='lon', elev_column='elev',
time_column='time', value_column='value')
fig, ax = pdb.plot() # plot to have a check
cfr.ProxyDatabase => a netCDF file#
Note that converting a cfr.ProxyDatabase to a netCDF file comes with the limitation that the time axis prior to 1 CE will be truncated since that is not supported yet.
[14]:
pdb.to_nc('./data/PAGES2k.nc')
100%|██████████| 692/692 [00:12<00:00, 54.30it/s]
ProxyDatabase saved to: ./data/PAGES2k.nc
[15]:
ds = xr.open_dataset('./data/PAGES2k.nc')
ds
/Users/fengzhu/Apps/miniconda3/envs/cfr-env/lib/python3.9/site-packages/xarray/coding/times.py:710: SerializationWarning: Unable to decode time axis into full numpy.datetime64 objects, continuing using cftime.datetime objects instead, reason: dates out of range
dtype = _decode_cf_datetime_dtype(data, units, calendar, self.use_cftime)
/Users/fengzhu/Apps/miniconda3/envs/cfr-env/lib/python3.9/site-packages/xarray/core/indexing.py:524: SerializationWarning: Unable to decode time axis into full numpy.datetime64 objects, continuing using cftime.datetime objects instead, reason: dates out of range
return np.asarray(array[self.key], dtype=None)
[15]:
<xarray.Dataset>
Dimensions: (time: 1156)
Coordinates:
* time (time) object 0850-01-01 00:00:00 ... 2005-01-01 00:00:00
Data variables: (12/692)
NAm_153 (time) float64 ...
Asi_245 (time) float64 ...
NAm_165 (time) float64 ...
Asi_178 (time) float64 ...
Asi_174 (time) float64 ...
Eur_016 (time) float64 ...
... ...
Ocn_169 (time) float64 ...
Asi_201 (time) float64 ...
Asi_179 (time) float64 ...
Arc_014 (time) float64 ...
Ocn_071 (time) float64 ...
Ocn_072 (time) float64 ...- time: 1156
- time(time)object0850-01-01 00:00:00 ... 2005-01-...
array([cftime.DatetimeGregorian(850, 1, 1, 0, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(851, 1, 1, 0, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(852, 1, 1, 0, 0, 0, 0, has_year_zero=False), ..., cftime.DatetimeGregorian(2003, 1, 1, 0, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(2004, 1, 1, 0, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(2005, 1, 1, 0, 0, 0, 0, has_year_zero=False)], dtype=object)
- NAm_153(time)float64...
- lat :
- 52.7
- lon :
- 241.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_245(time)float64...
- lat :
- 23.0
- lon :
- 114.0
- elev :
- nan
- ptype :
- documents
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_165(time)float64...
- lat :
- 37.9
- lon :
- 252.3
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_178(time)float64...
- lat :
- 28.77
- lon :
- 83.73
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_174(time)float64...
- lat :
- 28.18
- lon :
- 85.43
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Eur_016(time)float64...
- lat :
- 46.15
- lon :
- 7.5
- elev :
- nan
- ptype :
- lake.midge
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_198(time)float64...
- lat :
- 35.35
- lon :
- 71.93
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_145(time)float64...
- lat :
- 59.9
- lon :
- 223.2
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_070(time)float64...
- lat :
- 67.0
- lon :
- 309.3
- elev :
- nan
- ptype :
- lake.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_071(time)float64...
- lat :
- 68.49166667
- lon :
- 27.33333333
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_102(time)float64...
- lat :
- 62.0
- lon :
- 218.0
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_046(time)float64...
- lat :
- 46.0
- lon :
- 246.6
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_065(time)float64...
- lat :
- 25.84
- lon :
- 281.38
- elev :
- nan
- ptype :
- coral.calc
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_242(time)float64...
- lat :
- 30.5
- lon :
- 114.5
- elev :
- nan
- ptype :
- documents
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_170(time)float64...
- lat :
- 28.38
- lon :
- 83.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_023(time)float64...
- lat :
- -77.78
- lon :
- 158.72
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_045(time)float64...
- lat :
- 34.73
- lon :
- 100.78
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_149(time)float64...
- lat :
- 60.1
- lon :
- 225.6
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_193(time)float64...
- lat :
- 48.7
- lon :
- 241.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_194(time)float64...
- lat :
- 48.7
- lon :
- 241.7
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_089(time)float64...
- lat :
- 63.3
- lon :
- 212.2
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_042(time)float64...
- lat :
- 66.07
- lon :
- 214.6
- elev :
- nan
- ptype :
- lake.midge
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_188(time)float64...
- lat :
- 49.8
- lon :
- 245.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_184(time)float64...
- lat :
- 35.33
- lon :
- 74.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_214(time)float64...
- lat :
- 35.35
- lon :
- 71.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_075(time)float64...
- lat :
- -23.15
- lon :
- 43.58
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_147(time)float64...
- lat :
- 49.92
- lon :
- 91.57
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_098(time)float64...
- lat :
- 36.68
- lon :
- 98.42
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_099(time)float64...
- lat :
- 56.0
- lon :
- 228.0
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Eur_015(time)float64...
- lat :
- 46.5
- lon :
- 9.8
- elev :
- nan
- ptype :
- lake.reflectance
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_018(time)float64...
- lat :
- 45.89
- lon :
- 297.2
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_070(time)float64...
- lat :
- 28.9
- lon :
- 99.75
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_239(time)float64...
- lat :
- 28.0
- lon :
- 116.5
- elev :
- nan
- ptype :
- documents
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_011(time)float64...
- lat :
- 10.77
- lon :
- 295.23
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_215(time)float64...
- lat :
- 35.88
- lon :
- 74.88
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_052(time)float64...
- lat :
- 20.75
- lon :
- 341.4167
- elev :
- nan
- ptype :
- marine.foram
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_223(time)float64...
- lat :
- 29.62
- lon :
- 94.67
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_156(time)float64...
- lat :
- 52.6
- lon :
- 242.1
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_009(time)float64...
- lat :
- 50.48
- lon :
- 87.67
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_173(time)float64...
- lat :
- 29.52
- lon :
- 82.03
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_047(time)float64...
- lat :
- 37.93
- lon :
- 101.53
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_073(time)float64...
- lat :
- 46.2
- lon :
- 270.3
- elev :
- nan
- ptype :
- lake.pollen
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_078(time)float64...
- lat :
- -3.2
- lon :
- 40.1
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_154(time)float64...
- lat :
- 51.7
- lon :
- 243.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_148(time)float64...
- lat :
- 48.17
- lon :
- 99.87
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_191(time)float64...
- lat :
- 35.68
- lon :
- 71.63
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_062(time)float64...
- lat :
- 34.48
- lon :
- 110.08
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_019(time)float64...
- lat :
- 50.15
- lon :
- 85.37
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_167(time)float64...
- lat :
- 16.85
- lon :
- 112.33
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_085(time)float64...
- lat :
- 61.8
- lon :
- 212.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_086(time)float64...
- lat :
- 61.8
- lon :
- 212.7
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_091(time)float64...
- lat :
- -16.8167
- lon :
- 179.2333
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_093(time)float64...
- lat :
- -16.8167
- lon :
- 179.2333
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_155(time)float64...
- lat :
- 48.3
- lon :
- 98.93
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_045(time)float64...
- lat :
- 62.08
- lon :
- 342.18
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_024(time)float64...
- lat :
- -86.5
- lon :
- 252.01
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_098(time)float64...
- lat :
- 58.4
- lon :
- 225.4
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_004(time)float64...
- lat :
- -5.2012
- lon :
- 117.4867
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_073(time)float64...
- lat :
- 27.33
- lon :
- 99.3
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_209(time)float64...
- lat :
- 36.15
- lon :
- 74.18
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_094(time)float64...
- lat :
- 37.32
- lon :
- 98.4
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_001(time)float64...
- lat :
- 68.09
- lon :
- 209.53
- elev :
- nan
- ptype :
- lake.varve_thickness
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_096(time)float64...
- lat :
- -21.2378
- lon :
- 200.1722
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_068(time)float64...
- lat :
- 46.3
- lon :
- 292.1
- elev :
- nan
- ptype :
- lake.pollen
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_052(time)float64...
- lat :
- 34.78
- lon :
- 99.78
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_086(time)float64...
- lat :
- -1.5
- lon :
- 124.833
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_122(time)float64...
- lat :
- 51.8
- lon :
- 243.8
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_150(time)float64...
- lat :
- 48.7
- lon :
- 88.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_053(time)float64...
- lat :
- 38.556
- lon :
- 350.6502
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_117(time)float64...
- lat :
- 30.37
- lon :
- 130.53
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_186(time)float64...
- lat :
- 49.3
- lon :
- 245.6
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_161(time)float64...
- lat :
- 52.9
- lon :
- 242.0
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_030(time)float64...
- lat :
- 27.63
- lon :
- 90.13
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_101(time)float64...
- lat :
- -22.475
- lon :
- 166.4667
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_047(time)float64...
- lat :
- 34.23
- lon :
- 239.98000000000002
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_066(time)float64...
- lat :
- 46.7
- lon :
- 245.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_108(time)float64...
- lat :
- 43.77
- lon :
- 142.55
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_070(time)float64...
- lat :
- 24.6
- lon :
- 277.7
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_084(time)float64...
- lat :
- 37.47
- lon :
- 97.23
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_076(time)float64...
- lat :
- -3.2
- lon :
- 40.1
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_050(time)float64...
- lat :
- 38.5
- lon :
- 245.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_135(time)float64...
- lat :
- 42.2
- lon :
- 79.05
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_008(time)float64...
- lat :
- 30.85
- lon :
- 349.7315
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_144(time)float64...
- lat :
- 48.27
- lon :
- 88.87
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_185(time)float64...
- lat :
- 35.33
- lon :
- 74.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_029(time)float64...
- lat :
- 27.67
- lon :
- 90.72
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_115(time)float64...
- lat :
- 43.5
- lon :
- 143.2
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_133(time)float64...
- lat :
- 50.2
- lon :
- 242.9
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_134(time)float64...
- lat :
- 50.2
- lon :
- 242.9
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_243(time)float64...
- lat :
- 33.9167
- lon :
- 89.0833
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_059(time)float64...
- lat :
- 35.986
- lon :
- 355.2504
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_146(time)float64...
- lat :
- 49.37
- lon :
- 94.88
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_123(time)float64...
- lat :
- 49.4
- lon :
- 236.9
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_012(time)float64...
- lat :
- -79.57
- lon :
- 314.28
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_176(time)float64...
- lat :
- 27.67
- lon :
- 87.2
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_093(time)float64...
- lat :
- 65.2
- lon :
- 197.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_005(time)float64...
- lat :
- 36.2053
- lon :
- 355.6867
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_099(time)float64...
- lat :
- -17.5
- lon :
- 210.1667
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_128(time)float64...
- lat :
- 40.17
- lon :
- 72.62
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_228(time)float64...
- lat :
- 21.67
- lon :
- 104.1
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_020(time)float64...
- lat :
- -75.58
- lon :
- 356.57
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_090(time)float64...
- lat :
- 65.2
- lon :
- 197.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_144(time)float64...
- lat :
- 30.0867
- lon :
- 295.4583
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_063(time)float64...
- lat :
- 34.973
- lon :
- 284.799
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_202(time)float64...
- lat :
- 36.58
- lon :
- 75.08
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_189(time)float64...
- lat :
- 48.7
- lon :
- 239.3
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_171(time)float64...
- lat :
- 40.6
- lon :
- 254.2
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_178(time)float64...
- lat :
- -21.2378
- lon :
- 200.1722
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_203(time)float64...
- lat :
- 36.58
- lon :
- 75.08
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_029(time)float64...
- lat :
- 80.0
- lon :
- 318.86
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_054(time)float64...
- lat :
- -5.0
- lon :
- 133.44
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_113(time)float64...
- lat :
- 62.7
- lon :
- 249.0
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_020(time)float64...
- lat :
- 50.15
- lon :
- 85.37
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Eur_011(time)float64...
- lat :
- 49.0
- lon :
- 13.0
- elev :
- nan
- ptype :
- documents
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_171(time)float64...
- lat :
- 27.73
- lon :
- 86.33
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_158(time)float64...
- lat :
- -28.4667
- lon :
- 113.7667
- elev :
- nan
- ptype :
- coral.calc
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_157(time)float64...
- lat :
- 50.6
- lon :
- 245.0
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_124(time)float64...
- lat :
- 49.2
- lon :
- 234.8
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_006(time)float64...
- lat :
- 49.72
- lon :
- 87.28
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_033(time)float64...
- lat :
- 27.25
- lon :
- 89.38
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_149(time)float64...
- lat :
- 18.4667
- lon :
- 282.05
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_150(time)float64...
- lat :
- 18.4667
- lon :
- 282.05
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_120(time)float64...
- lat :
- 33.73
- lon :
- 133.12
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_035(time)float64...
- lat :
- 65.18
- lon :
- 316.17
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_001(time)float64...
- lat :
- -72.8
- lon :
- 159.06
- elev :
- nan
- ptype :
- ice.dD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_065(time)float64...
- lat :
- 30.33
- lon :
- 119.43
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_034(time)float64...
- lat :
- 1.4033
- lon :
- 119.078
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_049(time)float64...
- lat :
- 40.2
- lon :
- 244.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_050(time)float64...
- lat :
- 60.28
- lon :
- 25.42
- elev :
- nan
- ptype :
- lake.midge
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_091(time)float64...
- lat :
- 65.2
- lon :
- 197.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_177(time)float64...
- lat :
- 1.6792
- lon :
- 202.7527
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_035(time)float64...
- lat :
- -3.53
- lon :
- 119.2
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_080(time)float64...
- lat :
- 27.85
- lon :
- 34.32
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_171(time)float64...
- lat :
- -17.2667
- lon :
- 119.3667
- elev :
- nan
- ptype :
- coral.calc
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_152(time)float64...
- lat :
- 47.77
- lon :
- 107.0
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_043(time)float64...
- lat :
- 34.75
- lon :
- 100.82
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_049(time)float64...
- lat :
- 8.73
- lon :
- 109.869
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_108(time)float64...
- lat :
- -15.0
- lon :
- 166.99
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_028(time)float64...
- lat :
- 27.7
- lon :
- 90.68
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_025(time)float64...
- lat :
- 27.58
- lon :
- 90.65
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_103(time)float64...
- lat :
- 5.87
- lon :
- 197.87
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_027(time)float64...
- lat :
- -79.463
- lon :
- 247.875
- elev :
- nan
- ptype :
- borehole
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_109(time)float64...
- lat :
- 37.9
- lon :
- 240.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_079(time)float64...
- lat :
- 66.8
- lon :
- 68.0
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_060(time)float64...
- lat :
- 34.78
- lon :
- 100.82
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_110(time)float64...
- lat :
- 43.88
- lon :
- 145.6
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_029(time)float64...
- lat :
- 36.03
- lon :
- 141.78
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_007(time)float64...
- lat :
- 50.14
- lon :
- 87.72
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_087(time)float64...
- lat :
- 37.43
- lon :
- 98.05
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_036(time)float64...
- lat :
- 40.5
- lon :
- 4.03
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_116(time)float64...
- lat :
- 44.95
- lon :
- 142.12
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Eur_012(time)float64...
- lat :
- 42.5
- lon :
- 0.75
- elev :
- nan
- ptype :
- lake.chrysophyte
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Eur_004(time)float64...
- lat :
- 49.0
- lon :
- 20.0
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_068(time)float64...
- lat :
- 32.467
- lon :
- 295.3
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_069(time)float64...
- lat :
- 32.467
- lon :
- 295.3
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_075(time)float64...
- lat :
- 35.0
- lon :
- 100.07
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_093(time)float64...
- lat :
- 33.8
- lon :
- 96.13
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_043(time)float64...
- lat :
- 67.0
- lon :
- 309.3
- elev :
- nan
- ptype :
- lake.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_006(time)float64...
- lat :
- -79.46
- lon :
- 247.91
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Eur_020(time)float64...
- lat :
- 59.4
- lon :
- 24.75
- elev :
- nan
- ptype :
- documents
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_040(time)float64...
- lat :
- 29.28
- lon :
- 100.08
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_183(time)float64...
- lat :
- 49.0
- lon :
- 246.0
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_012(time)float64...
- lat :
- 50.48
- lon :
- 87.65
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_022(time)float64...
- lat :
- -72.82
- lon :
- 159.18
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_158(time)float64...
- lat :
- 52.2
- lon :
- 242.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Aus_002(time)float64...
- lat :
- -43.23
- lon :
- 170.28
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_133(time)float64...
- lat :
- 55.0
- lon :
- 160.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_078(time)float64...
- lat :
- 35.07
- lon :
- 100.35
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_145(time)float64...
- lat :
- 30.0867
- lon :
- 295.4583
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_019(time)float64...
- lat :
- 36.4
- lon :
- 241.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_098(time)float64...
- lat :
- 1.0
- lon :
- 173.0
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_025(time)float64...
- lat :
- 2.5
- lon :
- 9.38
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_002(time)float64...
- lat :
- -66.77
- lon :
- 112.807
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_018(time)float64...
- lat :
- -66.77
- lon :
- 112.807
- elev :
- nan
- ptype :
- borehole
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_151(time)float64...
- lat :
- 17.5333
- lon :
- 281.05
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_152(time)float64...
- lat :
- 17.5333
- lon :
- 281.05
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_114(time)float64...
- lat :
- 56.2
- lon :
- 292.3
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_115(time)float64...
- lat :
- 56.2
- lon :
- 292.3
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_042(time)float64...
- lat :
- 27.33
- lon :
- 99.3
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_103(time)float64...
- lat :
- 67.0
- lon :
- 208.0
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_080(time)float64...
- lat :
- 80.783
- lon :
- 65.63
- elev :
- nan
- ptype :
- ice.melt
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_078(time)float64...
- lat :
- 80.783
- lon :
- 65.63
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_115(time)float64...
- lat :
- 29.42
- lon :
- 34.97
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_009(time)float64...
- lat :
- 36.3
- lon :
- 241.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_125(time)float64...
- lat :
- 40.17
- lon :
- 72.58
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_017(time)float64...
- lat :
- 16.8402
- lon :
- 343.2673
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_034(time)float64...
- lat :
- 71.12
- lon :
- 322.68
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Eur_013(time)float64...
- lat :
- 62.0
- lon :
- 28.325
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_069(time)float64...
- lat :
- 34.63
- lon :
- 104.47
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_124(time)float64...
- lat :
- 40.17
- lon :
- 72.58
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_196(time)float64...
- lat :
- 36.33
- lon :
- 74.03
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_036(time)float64...
- lat :
- 72.58
- lon :
- 322.36
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_232(time)float64...
- lat :
- 28.38
- lon :
- 85.72
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_246(time)float64...
- lat :
- 25.0
- lon :
- 118.0
- elev :
- nan
- ptype :
- documents
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_129(time)float64...
- lat :
- 55.3
- lon :
- 282.2
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_130(time)float64...
- lat :
- 55.3
- lon :
- 282.2
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_104(time)float64...
- lat :
- 68.7
- lon :
- 218.4
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_204(time)float64...
- lat :
- 36.58
- lon :
- 75.08
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_110(time)float64...
- lat :
- 34.1
- lon :
- 243.2
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_061(time)float64...
- lat :
- 66.9
- lon :
- 65.6
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_170(time)float64...
- lat :
- -21.8333
- lon :
- 114.1833
- elev :
- nan
- ptype :
- coral.calc
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_180(time)float64...
- lat :
- 7.2859
- lon :
- 134.2503
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_154(time)float64...
- lat :
- -28.4589
- lon :
- 113.749
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_155(time)float64...
- lat :
- -28.4589
- lon :
- 113.749
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_090(time)float64...
- lat :
- 34.47
- lon :
- 110.08
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_187(time)float64...
- lat :
- 36.03
- lon :
- 74.58
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_008(time)float64...
- lat :
- 67.9
- lon :
- 219.3
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_026(time)float64...
- lat :
- 61.81
- lon :
- 24.68
- elev :
- nan
- ptype :
- lake.varve_property
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_013(time)float64...
- lat :
- 37.8
- lon :
- 240.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_059(time)float64...
- lat :
- 71.27
- lon :
- 333.27
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_011(time)float64...
- lat :
- 37.2
- lon :
- 241.9
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_089(time)float64...
- lat :
- 37.52
- lon :
- 97.05
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_175(time)float64...
- lat :
- 44.8
- lon :
- 289.2
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_135(time)float64...
- lat :
- 61.48
- lon :
- 340.47
- elev :
- nan
- ptype :
- marine.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_029(time)float64...
- lat :
- 52.7
- lon :
- 241.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_076(time)float64...
- lat :
- 45.89
- lon :
- 264.522
- elev :
- nan
- ptype :
- lake.pollen
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_077(time)float64...
- lat :
- 45.89
- lon :
- 264.522
- elev :
- nan
- ptype :
- lake.pollen
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_157(time)float64...
- lat :
- 47.27
- lon :
- 100.03
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_144(time)float64...
- lat :
- 58.0
- lon :
- 266.0
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_164(time)float64...
- lat :
- 46.52
- lon :
- 100.95
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_183(time)float64...
- lat :
- 35.17
- lon :
- 75.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_175(time)float64...
- lat :
- 27.78
- lon :
- 87.27
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_088(time)float64...
- lat :
- 37.45
- lon :
- 97.78
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_086(time)float64...
- lat :
- 37.45
- lon :
- 97.53
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_096(time)float64...
- lat :
- 37.03
- lon :
- 98.67
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_111(time)float64...
- lat :
- 17.93
- lon :
- 293.0
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_112(time)float64...
- lat :
- 17.93
- lon :
- 293.0
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Eur_014(time)float64...
- lat :
- 46.5
- lon :
- 9.8
- elev :
- nan
- ptype :
- lake.chironomid
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_180(time)float64...
- lat :
- 45.9
- lon :
- 245.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_181(time)float64...
- lat :
- 45.9
- lon :
- 245.7
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_088(time)float64...
- lat :
- 60.0
- lon :
- 218.3
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_154(time)float64...
- lat :
- 49.87
- lon :
- 91.43
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_116(time)float64...
- lat :
- 29.42
- lon :
- 34.97
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_046(time)float64...
- lat :
- 10.98
- lon :
- 74.9993
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_210(time)float64...
- lat :
- 35.17
- lon :
- 75.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_072(time)float64...
- lat :
- 27.37
- lon :
- 99.37
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_028(time)float64...
- lat :
- 76.62
- lon :
- 323.6
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_132(time)float64...
- lat :
- 24.8333
- lon :
- 65.9167
- elev :
- nan
- ptype :
- marine.foram
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_113(time)float64...
- lat :
- 43.22
- lon :
- 145.47
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_019(time)float64...
- lat :
- 43.53
- lon :
- 297.52
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_233(time)float64...
- lat :
- 30.0
- lon :
- 117.5
- elev :
- nan
- ptype :
- documents
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_182(time)float64...
- lat :
- 27.73
- lon :
- 87.2
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_056(time)float64...
- lat :
- -64.87
- lon :
- 295.8
- elev :
- nan
- ptype :
- marine.TEX86
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_130(time)float64...
- lat :
- -4.1916
- lon :
- 151.9772
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_131(time)float64...
- lat :
- -4.1916
- lon :
- 151.9772
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_159(time)float64...
- lat :
- 50.9
- lon :
- 244.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_147(time)float64...
- lat :
- 11.77
- lon :
- 293.25
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_094(time)float64...
- lat :
- 62.4
- lon :
- 216.9
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_095(time)float64...
- lat :
- 62.4
- lon :
- 216.9
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_151(time)float64...
- lat :
- 46.82
- lon :
- 100.12
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_009(time)float64...
- lat :
- 30.845
- lon :
- 349.9017
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_225(time)float64...
- lat :
- 29.35
- lon :
- 92.0
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_182(time)float64...
- lat :
- 46.0
- lon :
- 246.6
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_064(time)float64...
- lat :
- 80.52
- lon :
- 94.82
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_161(time)float64...
- lat :
- 48.25
- lon :
- 97.4
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_139(time)float64...
- lat :
- 48.6
- lon :
- 88.37
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_106(time)float64...
- lat :
- -16.82
- lon :
- 179.23
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_069(time)float64...
- lat :
- 45.3
- lon :
- 268.5
- elev :
- nan
- ptype :
- lake.pollen
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_092(time)float64...
- lat :
- 65.2
- lon :
- 197.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_030(time)float64...
- lat :
- 69.87
- lon :
- 291.17
- elev :
- nan
- ptype :
- lake.varve_thickness
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_174(time)float64...
- lat :
- -21.9
- lon :
- 113.9667
- elev :
- nan
- ptype :
- coral.calc
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_069(time)float64...
- lat :
- 78.0217
- lon :
- 13.9311
- elev :
- nan
- ptype :
- lake.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_199(time)float64...
- lat :
- 35.4
- lon :
- 74.12
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_222(time)float64...
- lat :
- 29.07
- lon :
- 93.95
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_077(time)float64...
- lat :
- -8.0167
- lon :
- 39.5
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_237(time)float64...
- lat :
- 23.5
- lon :
- 112.5
- elev :
- nan
- ptype :
- documents
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_046(time)float64...
- lat :
- 34.72
- lon :
- 100.72
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_075(time)float64...
- lat :
- 78.4
- lon :
- 279.6
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_106(time)float64...
- lat :
- 35.4
- lon :
- 249.8
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- SAm_031(time)float64...
- lat :
- -45.5167
- lon :
- 288.18330000000003
- elev :
- nan
- ptype :
- lake.BSi
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_129(time)float64...
- lat :
- 39.83
- lon :
- 71.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_116(time)float64...
- lat :
- 61.7
- lon :
- 220.3
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_163(time)float64...
- lat :
- 52.8
- lon :
- 242.3
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_083(time)float64...
- lat :
- -4.62
- lon :
- 55.0
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_097(time)float64...
- lat :
- 55.3
- lon :
- 228.4
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_022(time)float64...
- lat :
- 26.68
- lon :
- 266.07
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_169(time)float64...
- lat :
- 27.7
- lon :
- 86.28
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_039(time)float64...
- lat :
- 66.55
- lon :
- 342.58
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_160(time)float64...
- lat :
- 10.2773
- lon :
- 250.7869
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- SAm_026(time)float64...
- lat :
- -13.9333
- lon :
- 289.1667
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Aus_007(time)float64...
- lat :
- -42.27
- lon :
- 145.87
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_172(time)float64...
- lat :
- -23.0333
- lon :
- 113.8167
- elev :
- nan
- ptype :
- coral.calc
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Eur_017(time)float64...
- lat :
- 42.9
- lon :
- 356.5
- elev :
- nan
- ptype :
- speleothem.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_032(time)float64...
- lat :
- 60.2
- lon :
- 221.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_084(time)float64...
- lat :
- 63.7
- lon :
- 210.4
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_032(time)float64...
- lat :
- 27.42
- lon :
- 90.97
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_206(time)float64...
- lat :
- 35.5
- lon :
- 74.08
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_146(time)float64...
- lat :
- 59.8
- lon :
- 223.4
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_105(time)float64...
- lat :
- 57.1
- lon :
- 224.6
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_060(time)float64...
- lat :
- 25.38
- lon :
- 279.83
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_053(time)float64...
- lat :
- 67.25
- lon :
- 294.25
- elev :
- nan
- ptype :
- ice.melt
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_126(time)float64...
- lat :
- 67.2
- lon :
- 244.1
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_184(time)float64...
- lat :
- 43.0
- lon :
- 237.8
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Aus_004(time)float64...
- lat :
- -41.31
- lon :
- 147.75
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_037(time)float64...
- lat :
- 46.32
- lon :
- 152.53
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_135(time)float64...
- lat :
- 49.4
- lon :
- 242.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_075(time)float64...
- lat :
- 45.8
- lon :
- 284.9
- elev :
- nan
- ptype :
- lake.pollen
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_007(time)float64...
- lat :
- 32.784
- lon :
- 283.724
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_011(time)float64...
- lat :
- 72.1
- lon :
- 321.92
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_015(time)float64...
- lat :
- 50.48
- lon :
- 87.65
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_003(time)float64...
- lat :
- 50.87
- lon :
- 85.23
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_080(time)float64...
- lat :
- 45.3
- lon :
- 268.5
- elev :
- nan
- ptype :
- lake.pollen
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_021(time)float64...
- lat :
- 27.55
- lon :
- 266.07
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Afr_005(time)float64...
- lat :
- -10.0033
- lon :
- 34.2883
- elev :
- nan
- ptype :
- lake.TEX86
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_066(time)float64...
- lat :
- 65.9
- lon :
- 16.1
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_165(time)float64...
- lat :
- 46.78
- lon :
- 101.95
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_241(time)float64...
- lat :
- 32.1
- lon :
- 118.8
- elev :
- nan
- ptype :
- documents
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_083(time)float64...
- lat :
- 27.88
- lon :
- 98.4
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_010(time)float64...
- lat :
- -64.2017
- lon :
- 302.315
- elev :
- nan
- ptype :
- ice.dD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_122(time)float64...
- lat :
- -22.1
- lon :
- 153.0
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_197(time)float64...
- lat :
- 35.45
- lon :
- 74.78
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_080(time)float64...
- lat :
- 27.58
- lon :
- 99.28
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_166(time)float64...
- lat :
- -5.22
- lon :
- 145.82
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_023(time)float64...
- lat :
- 24.58
- lon :
- 280.74
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_156(time)float64...
- lat :
- 49.38
- lon :
- 94.88
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_092(time)float64...
- lat :
- 34.47
- lon :
- 110.08
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_095(time)float64...
- lat :
- 37.03
- lon :
- 98.63
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_123(time)float64...
- lat :
- -15.94
- lon :
- 166.04
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_140(time)float64...
- lat :
- 23.766
- lon :
- 283.947
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_141(time)float64...
- lat :
- 23.766
- lon :
- 283.947
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_054(time)float64...
- lat :
- 65.1
- lon :
- 276.21
- elev :
- nan
- ptype :
- lake.chironomid
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_195(time)float64...
- lat :
- 47.9
- lon :
- 236.2
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_121(time)float64...
- lat :
- 40.17
- lon :
- 72.58
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_190(time)float64...
- lat :
- 48.7
- lon :
- 239.3
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_011(time)float64...
- lat :
- -75.92
- lon :
- 275.75
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_196(time)float64...
- lat :
- 46.2
- lon :
- 238.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Eur_003(time)float64...
- lat :
- 68.0
- lon :
- 25.0
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_187(time)float64...
- lat :
- 37.6
- lon :
- 246.1
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_164(time)float64...
- lat :
- -17.73
- lon :
- 148.43
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_165(time)float64...
- lat :
- -17.73
- lon :
- 148.43
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_065(time)float64...
- lat :
- 44.5
- lon :
- 289.9
- elev :
- nan
- ptype :
- lake.pollen
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_172(time)float64...
- lat :
- 29.47
- lon :
- 82.12
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_179(time)float64...
- lat :
- 7.2708
- lon :
- 134.3837
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_141(time)float64...
- lat :
- 49.97
- lon :
- 91.0
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Eur_007(time)float64...
- lat :
- 46.4
- lon :
- 7.8
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_026(time)float64...
- lat :
- 51.4
- lon :
- 242.7
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_189(time)float64...
- lat :
- 36.03
- lon :
- 74.58
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Aus_005(time)float64...
- lat :
- -43.0
- lon :
- 171.0
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_137(time)float64...
- lat :
- 42.42
- lon :
- 78.97
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_082(time)float64...
- lat :
- 27.8483
- lon :
- 34.31
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_066(time)float64...
- lat :
- 30.6486
- lon :
- 295.01120000000003
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_067(time)float64...
- lat :
- 30.6486
- lon :
- 295.01120000000003
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_044(time)float64...
- lat :
- 75.33
- lon :
- 277.5
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_107(time)float64...
- lat :
- 31.37
- lon :
- 78.17
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_003(time)float64...
- lat :
- 66.97
- lon :
- 7.64
- elev :
- nan
- ptype :
- marine.diatom
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_063(time)float64...
- lat :
- 63.2475
- lon :
- 13.3375
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_138(time)float64...
- lat :
- 32.47
- lon :
- 295.3
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_136(time)float64...
- lat :
- 11.955
- lon :
- 44.3
- elev :
- nan
- ptype :
- marine.TEX86
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_137(time)float64...
- lat :
- 11.955
- lon :
- 44.3
- elev :
- nan
- ptype :
- marine.TEX86
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_180(time)float64...
- lat :
- 27.5
- lon :
- 87.98
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_007(time)float64...
- lat :
- -79.46
- lon :
- 247.91
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_028(time)float64...
- lat :
- -79.46
- lon :
- 247.91
- elev :
- nan
- ptype :
- ice.dD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_149(time)float64...
- lat :
- 48.5
- lon :
- 88.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_033(time)float64...
- lat :
- 80.7
- lon :
- 286.9
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_024(time)float64...
- lat :
- 50.15
- lon :
- 85.37
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_043(time)float64...
- lat :
- 27.2
- lon :
- 268.58
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_177(time)float64...
- lat :
- 27.73
- lon :
- 87.2
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_044(time)float64...
- lat :
- 45.3
- lon :
- 248.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- SAm_029(time)float64...
- lat :
- -41.17
- lon :
- 288.08
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_122(time)float64...
- lat :
- 40.17
- lon :
- 72.58
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_027(time)float64...
- lat :
- 73.94
- lon :
- 322.37
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_166(time)float64...
- lat :
- 47.95
- lon :
- 107.45
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Eur_005(time)float64...
- lat :
- 47.0
- lon :
- 25.3
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_168(time)float64...
- lat :
- 11.17
- lon :
- 299.15
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_138(time)float64...
- lat :
- 42.15
- lon :
- 79.45
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_045(time)float64...
- lat :
- 46.3
- lon :
- 246.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_038(time)float64...
- lat :
- 30.23
- lon :
- 100.27
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- SAm_024(time)float64...
- lat :
- -38.5
- lon :
- 288.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_039(time)float64...
- lat :
- 28.98
- lon :
- 99.93
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- SAm_030(time)float64...
- lat :
- -32.2667
- lon :
- 289.5
- elev :
- nan
- ptype :
- lake.reflectance
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_056(time)float64...
- lat :
- 34.78
- lon :
- 100.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_001(time)float64...
- lat :
- 66.55
- lon :
- 342.3
- elev :
- nan
- ptype :
- marine.diatom
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_048(time)float64...
- lat :
- 36.3
- lon :
- 98.08
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_032(time)float64...
- lat :
- 75.1
- lon :
- 317.68
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_126(time)float64...
- lat :
- 40.2
- lon :
- 72.58
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_053(time)float64...
- lat :
- 34.72
- lon :
- 99.67
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_142(time)float64...
- lat :
- 49.97
- lon :
- 90.98
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_205(time)float64...
- lat :
- 36.58
- lon :
- 75.08
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_016(time)float64...
- lat :
- 24.33
- lon :
- 276.74
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_020(time)float64...
- lat :
- 55.5
- lon :
- 346.1
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_194(time)float64...
- lat :
- 35.03
- lon :
- 74.58
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_127(time)float64...
- lat :
- 64.0
- lon :
- 256.1
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_140(time)float64...
- lat :
- 50.7
- lon :
- 238.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_141(time)float64...
- lat :
- 50.7
- lon :
- 238.5
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_096(time)float64...
- lat :
- 61.3
- lon :
- 217.3
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_142(time)float64...
- lat :
- 23.504
- lon :
- 283.423
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_143(time)float64...
- lat :
- 23.504
- lon :
- 283.423
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_088(time)float64...
- lat :
- 13.5982
- lon :
- 144.8359
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_077(time)float64...
- lat :
- 66.6
- lon :
- 17.6
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_129(time)float64...
- lat :
- 5.87
- lon :
- 197.87
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_110(time)float64...
- lat :
- 24.93
- lon :
- 279.25
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_051(time)float64...
- lat :
- 35.07
- lon :
- 100.35
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_105(time)float64...
- lat :
- 10.18
- lon :
- 76.87
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_002(time)float64...
- lat :
- 72.0
- lon :
- 101.0
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_219(time)float64...
- lat :
- 31.23
- lon :
- 96.48
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_077(time)float64...
- lat :
- 38.7
- lon :
- 99.68
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_065(time)float64...
- lat :
- 66.3
- lon :
- 18.2
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_001(time)float64...
- lat :
- 50.08
- lon :
- 87.77
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_007(time)float64...
- lat :
- 36.5
- lon :
- 241.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_067(time)float64...
- lat :
- 33.8
- lon :
- 281.0
- elev :
- nan
- ptype :
- lake.pollen
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Aus_009(time)float64...
- lat :
- -41.67
- lon :
- 145.65
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_013(time)float64...
- lat :
- -75.1
- lon :
- 123.39
- elev :
- nan
- ptype :
- ice.dD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_045(time)float64...
- lat :
- 67.25
- lon :
- 293.25
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_238(time)float64...
- lat :
- 35.28
- lon :
- 81.48
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_101(time)float64...
- lat :
- -5.5
- lon :
- 122.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_130(time)float64...
- lat :
- 39.83
- lon :
- 71.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_132(time)float64...
- lat :
- 39.83
- lon :
- 71.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_070(time)float64...
- lat :
- 43.0
- lon :
- 250.4
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_005(time)float64...
- lat :
- -75.0
- lon :
- 359.99
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_040(time)float64...
- lat :
- 61.43
- lon :
- 335.915
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_084(time)float64...
- lat :
- -2.5
- lon :
- 150.5
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_136(time)float64...
- lat :
- 42.15
- lon :
- 79.47
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_100(time)float64...
- lat :
- 33.72
- lon :
- 96.28
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_073(time)float64...
- lat :
- 20.83
- lon :
- 273.26
- elev :
- nan
- ptype :
- coral.calc
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_067(time)float64...
- lat :
- 61.5
- lon :
- 213.8
- elev :
- nan
- ptype :
- lake.BSi
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_014(time)float64...
- lat :
- -41.0
- lon :
- 285.55
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_114(time)float64...
- lat :
- -28.47
- lon :
- 113.77
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_051(time)float64...
- lat :
- 64.28
- lon :
- 30.12
- elev :
- nan
- ptype :
- lake.chironomid
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_082(time)float64...
- lat :
- 54.21
- lon :
- 288.65
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_163(time)float64...
- lat :
- 48.13
- lon :
- 100.27
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_081(time)float64...
- lat :
- -21.0333
- lon :
- 55.25
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_016(time)float64...
- lat :
- -77.32
- lon :
- 39.7
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_013(time)float64...
- lat :
- 50.48
- lon :
- 87.65
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_175(time)float64...
- lat :
- -16.7167
- lon :
- 146.0333
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_176(time)float64...
- lat :
- -16.7167
- lon :
- 146.0333
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_015(time)float64...
- lat :
- -77.32
- lon :
- 39.7
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_003(time)float64...
- lat :
- 60.5
- lon :
- 211.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_107(time)float64...
- lat :
- 32.5
- lon :
- 249.2
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_111(time)float64...
- lat :
- 30.33
- lon :
- 130.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Eur_019(time)float64...
- lat :
- 59.32
- lon :
- 18.06
- elev :
- nan
- ptype :
- documents
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_002(time)float64...
- lat :
- 49.62
- lon :
- 88.1
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_220(time)float64...
- lat :
- 29.3
- lon :
- 91.97
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_011(time)float64...
- lat :
- 50.48
- lon :
- 87.65
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_185(time)float64...
- lat :
- 45.3
- lon :
- 238.3
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_150(time)float64...
- lat :
- 51.1
- lon :
- 244.2
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_192(time)float64...
- lat :
- 46.2
- lon :
- 237.8
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_145(time)float64...
- lat :
- 48.35
- lon :
- 107.47
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_002(time)float64...
- lat :
- 63.76
- lon :
- 5.26
- elev :
- nan
- ptype :
- marine.foram
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_081(time)float64...
- lat :
- 28.03
- lon :
- 99.02
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_068(time)float64...
- lat :
- 33.72
- lon :
- 96.28
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_006(time)float64...
- lat :
- 24.83
- lon :
- 65.92
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_229(time)float64...
- lat :
- 12.22
- lon :
- 108.73
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_064(time)float64...
- lat :
- 6.3
- lon :
- 125.82
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_198(time)float64...
- lat :
- 41.3
- lon :
- 252.3
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_199(time)float64...
- lat :
- 41.3
- lon :
- 252.3
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_151(time)float64...
- lat :
- 52.2
- lon :
- 242.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_152(time)float64...
- lat :
- 52.2
- lon :
- 242.8
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_035(time)float64...
- lat :
- 27.95
- lon :
- 89.75
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_186(time)float64...
- lat :
- 36.03
- lon :
- 74.58
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_003(time)float64...
- lat :
- -84.0
- lon :
- 43.0
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_120(time)float64...
- lat :
- 52.3
- lon :
- 243.0
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_121(time)float64...
- lat :
- 52.3
- lon :
- 243.0
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_138(time)float64...
- lat :
- 49.7
- lon :
- 241.1
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_139(time)float64...
- lat :
- 49.7
- lon :
- 241.1
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_120(time)float64...
- lat :
- 27.1059
- lon :
- 142.1941
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_121(time)float64...
- lat :
- 27.1059
- lon :
- 142.1941
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_127(time)float64...
- lat :
- -21.2378
- lon :
- 200.1722
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_166(time)float64...
- lat :
- 39.3
- lon :
- 255.0
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_167(time)float64...
- lat :
- 39.3
- lon :
- 255.0
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_008(time)float64...
- lat :
- -79.3838
- lon :
- 248.76
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_128(time)float64...
- lat :
- -0.53296
- lon :
- 166.9283
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_158(time)float64...
- lat :
- 47.43
- lon :
- 100.42
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_060(time)float64...
- lat :
- 48.3
- lon :
- 239.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_074(time)float64...
- lat :
- 68.73
- lon :
- 15.73
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_013(time)float64...
- lat :
- 10.7
- lon :
- 295.06
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_085(time)float64...
- lat :
- 37.47
- lon :
- 97.22
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_017(time)float64...
- lat :
- -74.57
- lon :
- 273.1
- elev :
- nan
- ptype :
- ice.dD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_081(time)float64...
- lat :
- 36.5
- lon :
- 241.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_178(time)float64...
- lat :
- 45.8
- lon :
- 247.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_179(time)float64...
- lat :
- 45.8
- lon :
- 247.5
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_074(time)float64...
- lat :
- -8.2473
- lon :
- 115.5757
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_072(time)float64...
- lat :
- 78.8647
- lon :
- 17.425
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_142(time)float64...
- lat :
- 56.0
- lon :
- 299.0
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_083(time)float64...
- lat :
- 60.4
- lon :
- 212.2
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_235(time)float64...
- lat :
- 24.0
- lon :
- 121.0
- elev :
- nan
- ptype :
- documents
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_112(time)float64...
- lat :
- 68.6
- lon :
- 221.4
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_183(time)float64...
- lat :
- -18.315
- lon :
- 146.595
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_104(time)float64...
- lat :
- 31.2
- lon :
- 77.23
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_078(time)float64...
- lat :
- 45.3
- lon :
- 268.5
- elev :
- nan
- ptype :
- lake.pollen
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Eur_008(time)float64...
- lat :
- 44.0
- lon :
- 7.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_071(time)float64...
- lat :
- 31.78
- lon :
- 101.92
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_181(time)float64...
- lat :
- 27.83
- lon :
- 88.02
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_160(time)float64...
- lat :
- 53.0
- lon :
- 241.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_079(time)float64...
- lat :
- 27.58
- lon :
- 99.35
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_037(time)float64...
- lat :
- 27.45
- lon :
- 90.15
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_173(time)float64...
- lat :
- -17.5167
- lon :
- 118.9667
- elev :
- nan
- ptype :
- coral.calc
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_236(time)float64...
- lat :
- 23.16
- lon :
- 113.23
- elev :
- nan
- ptype :
- documents
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Eur_009(time)float64...
- lat :
- 42.5
- lon :
- 1.0
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_117(time)float64...
- lat :
- 54.9
- lon :
- 232.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_118(time)float64...
- lat :
- 54.9
- lon :
- 232.7
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_170(time)float64...
- lat :
- 39.9
- lon :
- 254.1
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_041(time)float64...
- lat :
- 39.9
- lon :
- 254.1
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_173(time)float64...
- lat :
- 44.6
- lon :
- 245.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_174(time)float64...
- lat :
- 44.6
- lon :
- 245.5
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_017(time)float64...
- lat :
- 50.48
- lon :
- 87.65
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_140(time)float64...
- lat :
- 46.32
- lon :
- 101.32
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_200(time)float64...
- lat :
- 35.83
- lon :
- 74.33
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_050(time)float64...
- lat :
- 57.45
- lon :
- 332.09
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_159(time)float64...
- lat :
- 49.7
- lon :
- 91.55
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_190(time)float64...
- lat :
- 35.68
- lon :
- 71.63
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_153(time)float64...
- lat :
- 48.83
- lon :
- 111.68
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_217(time)float64...
- lat :
- 31.95
- lon :
- 98.87
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_049(time)float64...
- lat :
- 37.0
- lon :
- 98.08
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_082(time)float64...
- lat :
- 28.03
- lon :
- 98.98
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_160(time)float64...
- lat :
- 48.98
- lon :
- 103.22
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_118(time)float64...
- lat :
- -21.905
- lon :
- 113.965
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_231(time)float64...
- lat :
- 42.17
- lon :
- 126.36
- elev :
- nan
- ptype :
- lake.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_005(time)float64...
- lat :
- 50.27
- lon :
- 87.83
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_027(time)float64...
- lat :
- 27.7
- lon :
- 90.77
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_195(time)float64...
- lat :
- 36.33
- lon :
- 74.03
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Aus_001(time)float64...
- lat :
- -41.83
- lon :
- 145.53
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_041(time)float64...
- lat :
- 24.8
- lon :
- 122.5
- elev :
- nan
- ptype :
- marine.TEX86
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_240(time)float64...
- lat :
- 43.955
- lon :
- 145.725
- elev :
- nan
- ptype :
- hybrid
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_139(time)float64...
- lat :
- -12.65
- lon :
- 45.1
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_061(time)float64...
- lat :
- -12.65
- lon :
- 45.1
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_062(time)float64...
- lat :
- -12.65
- lon :
- 45.1
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_197(time)float64...
- lat :
- 44.2
- lon :
- 252.9
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_168(time)float64...
- lat :
- 38.7
- lon :
- 252.4
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_169(time)float64...
- lat :
- 38.7
- lon :
- 252.4
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Eur_018(time)float64...
- lat :
- 47.1
- lon :
- 11.6
- elev :
- nan
- ptype :
- speleothem.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_071(time)float64...
- lat :
- 37.0
- lon :
- 243.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_119(time)float64...
- lat :
- 53.3
- lon :
- 239.3
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Aus_031(time)float64...
- lat :
- -40.07
- lon :
- 175.98
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_059(time)float64...
- lat :
- 34.78
- lon :
- 100.82
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_177(time)float64...
- lat :
- 25.1
- lon :
- 253.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_168(time)float64...
- lat :
- 27.7
- lon :
- 86.45
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_076(time)float64...
- lat :
- 27.62
- lon :
- 99.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_102(time)float64...
- lat :
- 30.98
- lon :
- 78.93
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_079(time)float64...
- lat :
- -0.13
- lon :
- 98.52
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_020(time)float64...
- lat :
- 82.83
- lon :
- 282.1
- elev :
- nan
- ptype :
- lake.varve_thickness
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_041(time)float64...
- lat :
- 29.15
- lon :
- 99.93
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_028(time)float64...
- lat :
- 32.977
- lon :
- 283.68399999999997
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_212(time)float64...
- lat :
- 35.17
- lon :
- 75.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_095(time)float64...
- lat :
- -16.8167
- lon :
- 179.2333
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_155(time)float64...
- lat :
- 52.9
- lon :
- 242.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_191(time)float64...
- lat :
- 48.7
- lon :
- 239.3
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_040(time)float64...
- lat :
- 61.35
- lon :
- 216.4
- elev :
- nan
- ptype :
- lake.midge
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_051(time)float64...
- lat :
- -44.15
- lon :
- 284.84000000000003
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_064(time)float64...
- lat :
- 44.4
- lon :
- 249.9
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_164(time)float64...
- lat :
- 40.1
- lon :
- 254.4
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_230(time)float64...
- lat :
- 29.0
- lon :
- 113.0
- elev :
- nan
- ptype :
- documents
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_057(time)float64...
- lat :
- 34.78
- lon :
- 100.82
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_055(time)float64...
- lat :
- 34.75
- lon :
- 99.68
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_022(time)float64...
- lat :
- 64.6
- lon :
- 340.2
- elev :
- nan
- ptype :
- lake.varve_thickness
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_156(time)float64...
- lat :
- -23.15
- lon :
- 43.58
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_004(time)float64...
- lat :
- 81.35
- lon :
- 290.47
- elev :
- nan
- ptype :
- lake.accumulation
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_061(time)float64...
- lat :
- 33.75
- lon :
- 107.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_027(time)float64...
- lat :
- -44.33
- lon :
- 287.03
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_131(time)float64...
- lat :
- 52.3
- lon :
- 242.7
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_058(time)float64...
- lat :
- 78.91
- lon :
- 6.77
- elev :
- nan
- ptype :
- marine.foram
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_048(time)float64...
- lat :
- -29.14
- lon :
- 16.72
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_079(time)float64...
- lat :
- 42.083
- lon :
- 236.584
- elev :
- nan
- ptype :
- speleothem.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_244(time)float64...
- lat :
- 55.0
- lon :
- 59.5
- elev :
- nan
- ptype :
- borehole
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_024(time)float64...
- lat :
- 24.765
- lon :
- 280.71
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_111(time)float64...
- lat :
- 58.4
- lon :
- 291.6
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_148(time)float64...
- lat :
- 59.6
- lon :
- 226.6
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_104(time)float64...
- lat :
- 7.95
- lon :
- 278.0
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_216(time)float64...
- lat :
- 51.83
- lon :
- 143.13
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_015(time)float64...
- lat :
- 24.59
- lon :
- 276.42
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_147(time)float64...
- lat :
- 61.4
- lon :
- 231.6
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_213(time)float64...
- lat :
- 35.0
- lon :
- 70.78
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_037(time)float64...
- lat :
- 64.77
- lon :
- 341.63
- elev :
- nan
- ptype :
- documents
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_008(time)float64...
- lat :
- 36.3
- lon :
- 241.6
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_099(time)float64...
- lat :
- 32.67
- lon :
- 95.72
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_125(time)float64...
- lat :
- 49.9
- lon :
- 241.1
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_107(time)float64...
- lat :
- 1.0
- lon :
- 172.0
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_091(time)float64...
- lat :
- 34.47
- lon :
- 110.08
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_005(time)float64...
- lat :
- 77.17
- lon :
- 298.87
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_068(time)float64...
- lat :
- 65.2
- lon :
- 15.5
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_128(time)float64...
- lat :
- 65.0
- lon :
- 221.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_087(time)float64...
- lat :
- 58.1
- lon :
- 207.3
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_019(time)float64...
- lat :
- -77.515
- lon :
- 167.6765
- elev :
- nan
- ptype :
- ice.dD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_162(time)float64...
- lat :
- 53.2
- lon :
- 240.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_058(time)float64...
- lat :
- 34.78
- lon :
- 100.82
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_192(time)float64...
- lat :
- 35.9
- lon :
- 71.73
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_010(time)float64...
- lat :
- 50.68
- lon :
- 87.97
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_032(time)float64...
- lat :
- 66.97
- lon :
- 7.63
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_022(time)float64...
- lat :
- 50.15
- lon :
- 85.37
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_157(time)float64...
- lat :
- -23.3572
- lon :
- 43.6195
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_176(time)float64...
- lat :
- 31.0
- lon :
- 244.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_112(time)float64...
- lat :
- 44.35
- lon :
- 142.18
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_188(time)float64...
- lat :
- 36.03
- lon :
- 74.58
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_036(time)float64...
- lat :
- 27.45
- lon :
- 90.15
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_050(time)float64...
- lat :
- 36.65
- lon :
- 98.08
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_066(time)float64...
- lat :
- 37.03
- lon :
- 98.68
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_021(time)float64...
- lat :
- 50.15
- lon :
- 85.37
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- SAm_006(time)float64...
- lat :
- -40.1
- lon :
- 287.95
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_059(time)float64...
- lat :
- 47.3
- lon :
- 238.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_001(time)float64...
- lat :
- 35.3
- lon :
- 248.6
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_162(time)float64...
- lat :
- 47.78
- lon :
- 107.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_030(time)float64...
- lat :
- 50.8
- lon :
- 244.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_123(time)float64...
- lat :
- 40.17
- lon :
- 72.58
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_146(time)float64...
- lat :
- 16.76
- lon :
- 337.1117
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_016(time)float64...
- lat :
- 50.48
- lon :
- 87.65
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- SAm_003(time)float64...
- lat :
- -33.85
- lon :
- 289.08
- elev :
- nan
- ptype :
- lake.reflectance
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Aus_029(time)float64...
- lat :
- -43.3646
- lon :
- 146.8749
- elev :
- nan
- ptype :
- lake.reflectance
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_109(time)float64...
- lat :
- 43.77
- lon :
- 142.55
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_014(time)float64...
- lat :
- -75.1
- lon :
- 123.39
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_074(time)float64...
- lat :
- 43.18
- lon :
- 87.18
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_055(time)float64...
- lat :
- 78.96
- lon :
- 5.885
- elev :
- nan
- ptype :
- marine.MAT
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_181(time)float64...
- lat :
- -15.7
- lon :
- 167.2
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_182(time)float64...
- lat :
- -15.7
- lon :
- 167.2
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_200(time)float64...
- lat :
- 44.8
- lon :
- 252.1
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_002(time)float64...
- lat :
- 67.1
- lon :
- 200.4
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_143(time)float64...
- lat :
- 47.1
- lon :
- 90.97
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_034(time)float64...
- lat :
- 27.45
- lon :
- 90.0
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_054(time)float64...
- lat :
- 34.75
- lon :
- 99.68
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_014(time)float64...
- lat :
- 50.48
- lon :
- 87.65
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_162(time)float64...
- lat :
- 10.2773
- lon :
- 250.7869
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_163(time)float64...
- lat :
- 10.2773
- lon :
- 250.7869
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_161(time)float64...
- lat :
- 10.2773
- lon :
- 250.7869
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_062(time)float64...
- lat :
- 68.26
- lon :
- 19.6
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_221(time)float64...
- lat :
- 31.12
- lon :
- 97.03
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_172(time)float64...
- lat :
- 43.9
- lon :
- 245.3
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_072(time)float64...
- lat :
- 50.2
- lon :
- 237.1
- elev :
- nan
- ptype :
- lake.varve_thickness
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_118(time)float64...
- lat :
- 30.37
- lon :
- 130.53
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_125(time)float64...
- lat :
- -21.2378
- lon :
- 200.1722
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_023(time)float64...
- lat :
- 50.15
- lon :
- 85.37
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_041(time)float64...
- lat :
- 61.9
- lon :
- 214.34
- elev :
- nan
- ptype :
- lake.midge
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- SAm_025(time)float64...
- lat :
- -39.33
- lon :
- 288.75
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_090(time)float64...
- lat :
- -4.15
- lon :
- 144.8833
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_067(time)float64...
- lat :
- 33.8
- lon :
- 96.13
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_148(time)float64...
- lat :
- 43.6561
- lon :
- 290.1983
- elev :
- nan
- ptype :
- bivalve.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_064(time)float64...
- lat :
- 43.85
- lon :
- 93.3
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_103(time)float64...
- lat :
- 33.08
- lon :
- 76.43
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_211(time)float64...
- lat :
- 35.17
- lon :
- 75.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_201(time)float64...
- lat :
- 43.7
- lon :
- 249.9
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_202(time)float64...
- lat :
- 43.7
- lon :
- 249.9
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_033(time)float64...
- lat :
- -7.4
- lon :
- 115.2
- elev :
- nan
- ptype :
- marine.MgCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_134(time)float64...
- lat :
- 43.35
- lon :
- 77.35
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Aus_030(time)float64...
- lat :
- -47.0
- lon :
- 167.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_226(time)float64...
- lat :
- 19.28
- lon :
- 98.93
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_063(time)float64...
- lat :
- 33.65
- lon :
- 107.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Eur_006(time)float64...
- lat :
- 47.0
- lon :
- 10.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_132(time)float64...
- lat :
- 56.9
- lon :
- 298.5
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_025(time)float64...
- lat :
- 66.73
- lon :
- 298.65
- elev :
- nan
- ptype :
- lake.varve_thickness
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_119(time)float64...
- lat :
- 10.2773
- lon :
- 250.7869
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_018(time)float64...
- lat :
- 50.15
- lon :
- 85.37
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_004(time)float64...
- lat :
- 50.3
- lon :
- 87.83
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_031(time)float64...
- lat :
- 27.58
- lon :
- 90.65
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Afr_012(time)float64...
- lat :
- -24.0
- lon :
- 29.18
- elev :
- nan
- ptype :
- speleothem.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_018(time)float64...
- lat :
- 79.83
- lon :
- 24.02
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_106(time)float64...
- lat :
- 10.0
- lon :
- 76.67
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_007(time)float64...
- lat :
- 61.03
- lon :
- 213.41
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_109(time)float64...
- lat :
- -19.9333
- lon :
- 185.2833
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_167(time)float64...
- lat :
- 29.48
- lon :
- 82.08
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_076(time)float64...
- lat :
- 62.917
- lon :
- 290.117
- elev :
- nan
- ptype :
- lake.varve_thickness
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_097(time)float64...
- lat :
- -5.2167
- lon :
- 145.8167
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_114(time)float64...
- lat :
- 35.73
- lon :
- 138.22
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_136(time)float64...
- lat :
- 50.6
- lon :
- 241.4
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_137(time)float64...
- lat :
- 50.6
- lon :
- 241.4
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_127(time)float64...
- lat :
- 39.92
- lon :
- 71.47
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_218(time)float64...
- lat :
- 31.12
- lon :
- 97.03
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_203(time)float64...
- lat :
- 41.4
- lon :
- 253.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_204(time)float64...
- lat :
- 41.4
- lon :
- 253.8
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_025(time)float64...
- lat :
- -77.3302
- lon :
- 162.5332
- elev :
- nan
- ptype :
- ice.dD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_207(time)float64...
- lat :
- 35.5
- lon :
- 74.75
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Afr_004(time)float64...
- lat :
- -6.03
- lon :
- 28.53
- elev :
- nan
- ptype :
- lake.TEX86
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_131(time)float64...
- lat :
- 40.17
- lon :
- 72.58
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_108(time)float64...
- lat :
- 37.8
- lon :
- 240.7
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_008(time)float64...
- lat :
- 50.12
- lon :
- 87.92
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_016(time)float64...
- lat :
- 69.5
- lon :
- 147.0
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_159(time)float64...
- lat :
- 10.2773
- lon :
- 250.7869
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_004(time)float64...
- lat :
- -70.86
- lon :
- 11.54
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_234(time)float64...
- lat :
- 34.0
- lon :
- 120.0
- elev :
- nan
- ptype :
- documents
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_193(time)float64...
- lat :
- 35.9
- lon :
- 71.73
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_133(time)float64...
- lat :
- 41.1
- lon :
- 351.1
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_087(time)float64...
- lat :
- -0.4
- lon :
- 268.77
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_100(time)float64...
- lat :
- 65.2
- lon :
- 197.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_101(time)float64...
- lat :
- 65.2
- lon :
- 197.7
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_030(time)float64...
- lat :
- 43.48
- lon :
- 305.13
- elev :
- nan
- ptype :
- marine.alkenone
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_024(time)float64...
- lat :
- 70.67
- lon :
- 125.87
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_021(time)float64...
- lat :
- -75.17
- lon :
- 6.5
- elev :
- nan
- ptype :
- ice.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_224(time)float64...
- lat :
- 30.3
- lon :
- 91.52
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_018(time)float64...
- lat :
- 36.3
- lon :
- 241.7
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_073(time)float64...
- lat :
- 68.625
- lon :
- 226.13
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_044(time)float64...
- lat :
- 34.73
- lon :
- 100.8
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_097(time)float64...
- lat :
- 36.75
- lon :
- 98.22
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ant_026(time)float64...
- lat :
- -78.2785
- lon :
- 104.8005
- elev :
- nan
- ptype :
- ice.dD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_227(time)float64...
- lat :
- 24.53
- lon :
- 121.38
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_143(time)float64...
- lat :
- 69.5
- lon :
- 232.2
- elev :
- nan
- ptype :
- tree.MXD
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_208(time)float64...
- lat :
- 36.15
- lon :
- 74.18
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_119(time)float64...
- lat :
- 30.33
- lon :
- 130.45
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_153(time)float64...
- lat :
- -28.4609
- lon :
- 113.772
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- NAm_074(time)float64...
- lat :
- 48.0
- lon :
- 269.0
- elev :
- nan
- ptype :
- lake.pollen
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_026(time)float64...
- lat :
- 27.67
- lon :
- 90.73
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_169(time)float64...
- lat :
- 11.17
- lon :
- 299.15
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_201(time)float64...
- lat :
- 35.88
- lon :
- 74.18
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Asi_179(time)float64...
- lat :
- 27.5
- lon :
- 88.02
- elev :
- nan
- ptype :
- tree.TRW
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Arc_014(time)float64...
- lat :
- 63.62
- lon :
- 29.1
- elev :
- nan
- ptype :
- lake.varve_thickness
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_071(time)float64...
- lat :
- 16.2
- lon :
- 298.51
- elev :
- nan
- ptype :
- coral.d18O
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- Ocn_072(time)float64...
- lat :
- 16.2
- lon :
- 298.51
- elev :
- nan
- ptype :
- coral.SrCa
- dt :
- 1.0
- time_name :
- Time
- time_unit :
- yr
- value_name :
- Proxy Value
- value_unit :
- none
[1156 values with dtype=float64]
- timePandasIndex
PandasIndex(CFTimeIndex([0850-01-01 00:00:00, 0851-01-01 00:00:00, 0852-01-01 00:00:00, 0853-01-01 00:00:00, 0854-01-01 00:00:00, 0855-01-01 00:00:00, 0856-01-01 00:00:00, 0857-01-01 00:00:00, 0858-01-01 00:00:00, 0859-01-01 00:00:00, ... 1996-01-01 00:00:00, 1997-01-01 00:00:00, 1998-01-01 00:00:00, 1999-01-01 00:00:00, 2000-01-01 00:00:00, 2001-01-01 00:00:00, 2002-01-01 00:00:00, 2003-01-01 00:00:00, 2004-01-01 00:00:00, 2005-01-01 00:00:00], dtype='object', length=1156, calendar='standard', freq='AS-JAN'))
a netCDF file => cfr.ProxyDatabase#
Now we load the generated netCDF file into a cfr.ProxyDatabase.
[17]:
pdb = cfr.ProxyDatabase().load_nc('./data/PAGES2k.nc')
fig, ax = pdb.plot() # plot to have a check
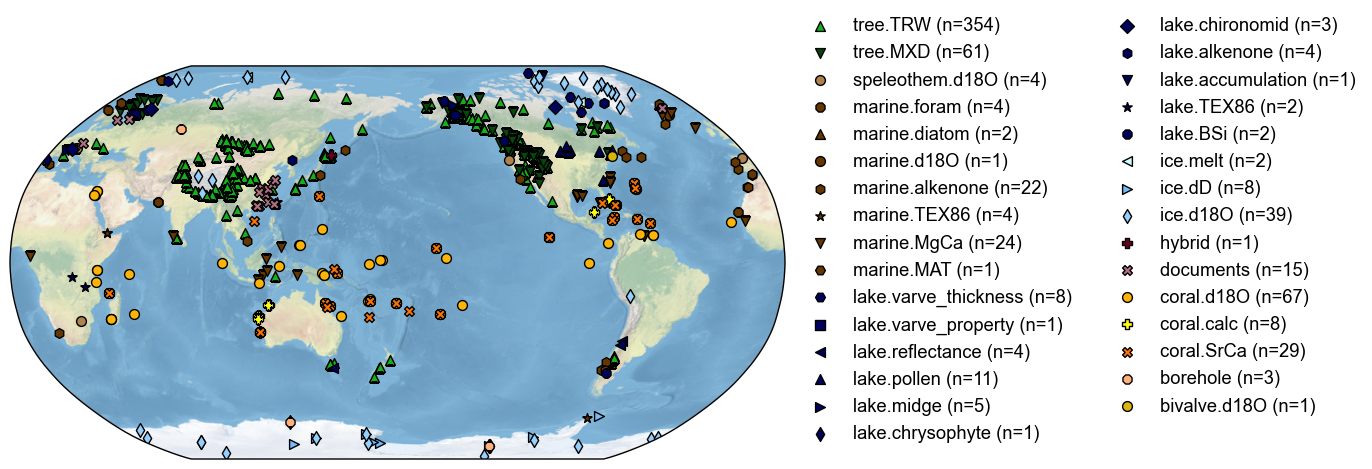
cfr.ProxyRecord => a netCDF file#
Each cfr.ProxyRecord can be saved to a netCDF file as well.
[18]:
pdb.records['NAm_153'].to_nc('./data/NAm_153.nc')
ProxyRecord saved to: ./data/NAm_153.nc
a netCDF file => cfr.ProxyRecord#
Now we load the saved netCDF file to a cfr.ProxyRecord.
[19]:
pobj = cfr.ProxyRecord().load_nc('./data/NAm_153.nc')
fig, ax = pobj.plot() # plot the record to have a check
/Users/fengzhu/Apps/miniconda3/envs/cfr-env/lib/python3.9/site-packages/xarray/coding/times.py:710: SerializationWarning: Unable to decode time axis into full numpy.datetime64 objects, continuing using cftime.datetime objects instead, reason: dates out of range
dtype = _decode_cf_datetime_dtype(data, units, calendar, self.use_cftime)
/Users/fengzhu/Apps/miniconda3/envs/cfr-env/lib/python3.9/site-packages/xarray/core/indexing.py:524: SerializationWarning: Unable to decode time axis into full numpy.datetime64 objects, continuing using cftime.datetime objects instead, reason: dates out of range
return np.asarray(array[self.key], dtype=None)
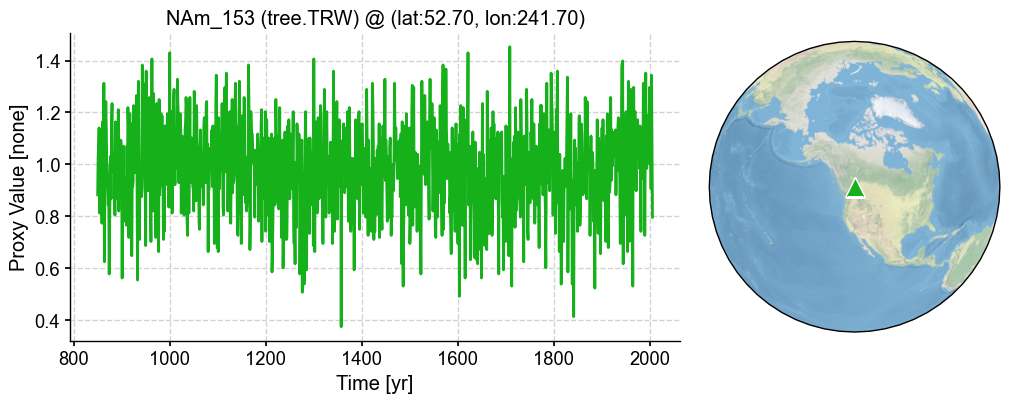
[ ]:
