Reconstructing the tropical Pacific SST with PAGES2k#
[1]:
%load_ext autoreload
%autoreload 2
import cfr
print(cfr.__version__)
2024.1.25
Load the PAGES2k database#
[2]:
job = cfr.ReconJob()
job.load_proxydb('PAGES2kv2')
Filter the database#
We only need the corals in this example.
[3]:
job.filter_proxydb(by='ptype', keys=['coral'])
fig, ax = job.proxydb.plot(plot_count=True)
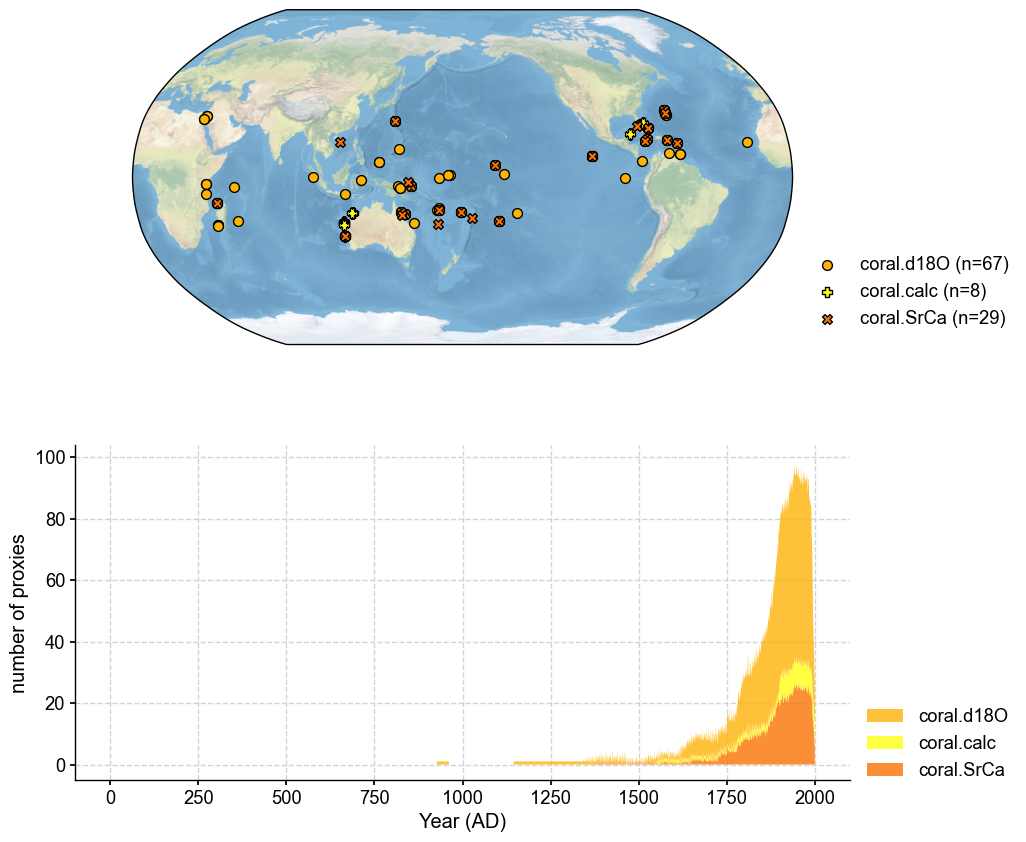
Annualize the database#
[4]:
job.annualize_proxydb(months=[12, 1, 2], ptypes=['coral'], verbose=True)
>>> job.configs["annualize_proxydb_months"] = [12, 1, 2]
>>> job.configs["annualize_proxydb_ptypes"] = ['coral']
Annualizing ProxyDatabase: 100%|██████████| 104/104 [00:01<00:00, 56.30it/s]
>>> 99 records remaining
>>> job.proxydb updated
Load the gridded climate data#
Load the model prior#
[5]:
job.load_clim(
tag='prior',
path_dict={
'tas': 'iCESM_past1000historical/tas',
},
anom_period=(1951, 1980),
)
>>> The target file seems existed at: ./data/tas_sfc_Amon_iCESM_past1000historical_085001-200512.nc . Loading from it instead of downloading ...
Load the instrumental observations#
[6]:
job.load_clim(
tag='obs',
path_dict={
'tas': 'gistemp1200_GHCNv4_ERSSTv5',
},
rename_dict={'tas': 'tempanomaly'},
anom_period=(1951, 1980),
)
>>> The target file seems existed at: ./data/gistemp1200_GHCNv4_ERSSTv5.nc.gz . Loading from it instead of downloading ...
Proxy system modeling#
Calibrating the proxy system models#
[7]:
ptype_psm_dict = {
'coral.d18O': 'Linear',
'coral.calc': 'Linear',
'coral.SrCa': 'Linear',
}
ptype_season_dict = {
'coral.d18O': [12, 1, 2],
'coral.calc': [12, 1, 2],
'coral.SrCa': [12, 1, 2],
}
ptype_clim_dict = {
'coral.d18O': ['tas'],
'coral.calc': ['tas'],
'coral.SrCa': ['tas'],
}
job.calib_psms(
ptype_psm_dict=ptype_psm_dict,
ptype_season_dict=ptype_season_dict,
ptype_clim_dict=ptype_clim_dict,
calib_period=(1850, 2015),
verbose=True,
)
>>> job.configs["ptype_psm_dict"] = {'coral.SrCa': 'Linear', 'coral.calc': 'Linear', 'coral.d18O': 'Linear'}
>>> job.configs["ptype_season_dict"] = {'coral.SrCa': [12, 1, 2], 'coral.calc': [12, 1, 2], 'coral.d18O': [12, 1, 2]}
>>> job.configs["psm_calib_period"] = (1850, 2015)
Calibrating the PSMs: 5%|▌ | 5/99 [00:00<00:02, 46.00it/s]
The number of overlapped data points is 14 < 25. Skipping ...
Calibrating the PSMs: 19%|█▉ | 19/99 [00:00<00:01, 61.99it/s]
The number of overlapped data points is 20 < 25. Skipping ...
The number of overlapped data points is 22 < 25. Skipping ...
The number of overlapped data points is 9 < 25. Skipping ...
Calibrating the PSMs: 32%|███▏ | 32/99 [00:00<00:01, 52.08it/s]
The number of overlapped data points is 24 < 25. Skipping ...
Calibrating the PSMs: 60%|█████▉ | 59/99 [00:01<00:00, 60.59it/s]
The number of overlapped data points is 22 < 25. Skipping ...
The number of overlapped data points is 22 < 25. Skipping ...
The number of overlapped data points is 0 < 25. Skipping ...
Calibrating the PSMs: 80%|███████▉ | 79/99 [00:01<00:00, 55.60it/s]
The number of overlapped data points is 21 < 25. Skipping ...
Calibrating the PSMs: 100%|██████████| 99/99 [00:01<00:00, 57.24it/s]
>>> PSM for Ocn_144 failed to be calibrated.
>>> PSM for Ocn_149 failed to be calibrated.
>>> PSM for Ocn_150 failed to be calibrated.
>>> PSM for Ocn_145 failed to be calibrated.
>>> PSM for Ocn_152 failed to be calibrated.
>>> PSM for Ocn_164 failed to be calibrated.
>>> PSM for Ocn_165 failed to be calibrated.
>>> PSM for Ocn_138 failed to be calibrated.
>>> PSM for Ocn_183 failed to be calibrated.
>>> 90 records tagged "calibrated" with ProxyRecord.psm created
Forwarding the proxy system models#
[8]:
job.forward_psms()
Forwarding the PSMs: 100%|██████████| 90/90 [00:07<00:00, 12.77it/s]
Annualizing, regridding, and cropping the prior field#
[9]:
job.annualize_clim(tag='prior', months=[12, 1, 2], verbose=True)
job.regrid_clim(tag='prior', nlat=42, nlon=63, verbose=True)
job.crop_clim(tag='prior', lat_min=-35, lat_max=35, verbose=True)
>>> job.configs["prior_annualize_months"] = [12, 1, 2]
>>> Processing tas ...
>>> job.prior updated
>>> job.configs["prior_regrid_nlat"] = 42
>>> job.configs["prior_regrid_nlon"] = 63
>>> Processing tas ...
>>> job.configs["prior_lat_min"] = -35
>>> job.configs["prior_lat_max"] = 35
>>> job.configs["prior_lon_min"] = 0
>>> job.configs["prior_lon_max"] = 360
>>> Processing tas ...
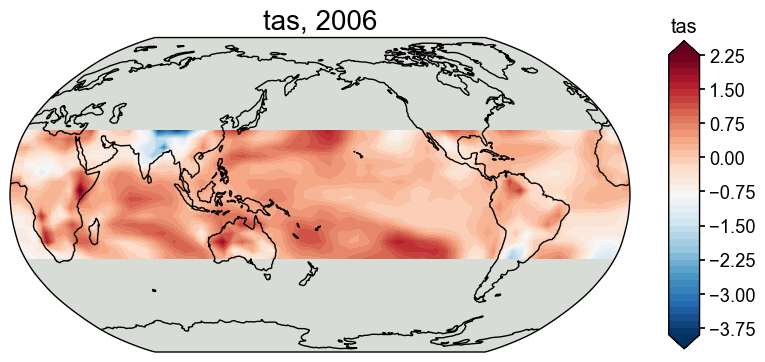
[10]:
fig, ax = job.prior['tas'][-1].plot()
Run the DA solver#
[11]:
job.run_da_mc(
save_dirpath='./recons/lmr-real-pages2k',
recon_seeds=list(range(1, 11)),
verbose=True,
)
>>> job.configs["recon_period"] = [0, 2000]
>>> job.configs["recon_loc_rad"] = 25000
>>> job.configs["recon_timescale"] = 1
>>> job.configs["recon_vars"] = ['tas']
>>> job.configs["nens"] = 100
>>> job.configs["recon_seeds"] = [1, 2, 3, 4, 5, 6, 7, 8, 9, 10]
>>> job.configs["assim_frac"] = 0.75
>>> job.configs["save_dirpath"] = ./recons/lmr-real-pages2k
>>> job.configs["compress_params"] = {'zlib': True}
>>> job.configs["output_full_ens"] = False
>>> job.configs["recon_sampling_mode"] = fixed
>>> job.configs["trim_prior"] = True
>>> job.configs["allownan"] = False
>>> seed: 1 | max: 10
KF updating: 100%|██████████| 2001/2001 [00:45<00:00, 43.92it/s]
>>> Reconstructed fields saved to: ./recons/lmr-real-pages2k/job_r01_recon.nc
>>> seed: 2 | max: 10
KF updating: 100%|██████████| 2001/2001 [00:51<00:00, 38.99it/s]
>>> Reconstructed fields saved to: ./recons/lmr-real-pages2k/job_r02_recon.nc
>>> seed: 3 | max: 10
KF updating: 100%|██████████| 2001/2001 [00:40<00:00, 49.28it/s]
>>> Reconstructed fields saved to: ./recons/lmr-real-pages2k/job_r03_recon.nc
>>> seed: 4 | max: 10
KF updating: 100%|██████████| 2001/2001 [00:41<00:00, 48.73it/s]
>>> Reconstructed fields saved to: ./recons/lmr-real-pages2k/job_r04_recon.nc
>>> seed: 5 | max: 10
KF updating: 100%|██████████| 2001/2001 [00:37<00:00, 53.29it/s]
>>> Reconstructed fields saved to: ./recons/lmr-real-pages2k/job_r05_recon.nc
>>> seed: 6 | max: 10
KF updating: 100%|██████████| 2001/2001 [00:40<00:00, 50.02it/s]
>>> Reconstructed fields saved to: ./recons/lmr-real-pages2k/job_r06_recon.nc
>>> seed: 7 | max: 10
KF updating: 100%|██████████| 2001/2001 [00:42<00:00, 47.15it/s]
>>> Reconstructed fields saved to: ./recons/lmr-real-pages2k/job_r07_recon.nc
>>> seed: 8 | max: 10
KF updating: 100%|██████████| 2001/2001 [00:21<00:00, 94.50it/s]
>>> Reconstructed fields saved to: ./recons/lmr-real-pages2k/job_r08_recon.nc
>>> seed: 9 | max: 10
KF updating: 100%|██████████| 2001/2001 [00:23<00:00, 84.33it/s]
>>> Reconstructed fields saved to: ./recons/lmr-real-pages2k/job_r09_recon.nc
>>> seed: 10 | max: 10
KF updating: 100%|██████████| 2001/2001 [00:23<00:00, 84.80it/s]
>>> Reconstructed fields saved to: ./recons/lmr-real-pages2k/job_r10_recon.nc
>>> DONE! Total time spent: 12.28 mins.
Validate the reconstruction#
[12]:
res = cfr.ReconRes('./recons/lmr-real-pages2k')
res.load(['nino3.4', 'tas'], verbose=True)
>>> ReconRes.recons["nino3.4"] created
>>> ReconRes.da["nino3.4"] created
>>> ReconRes.recons["tas"] created
>>> ReconRes.da["tas"] created
Load the validation target#
[13]:
target = cfr.ClimateField().fetch('HadCRUT4.6_GraphEM', vn='tas').get_anom((1951, 1980))
>>> The target file seems existed at: ./data/HadCRUT4.6_GraphEM_median.nc . Loading from it instead of downloading ...
[14]:
target = target.annualize(months=[12, 1, 2]).crop(lat_min=-35, lat_max=35)
target.da
[14]:
<xarray.DataArray 'tas' (time: 169, lat: 14, lon: 72)>
array([[[-0.8966176 , -1.00779957, -0.90773527, ..., -0.04003081,
0.10966071, -0.6124733 ],
[-0.42084288, -1.02308963, -0.54711297, ..., 0.90217373,
0.29498114, -0.07187287],
[-0.82761943, -0.62889297, -0.39431214, ..., -0.11959601,
-0.31819982, -0.72588533],
...,
[-1.7816816 , -1.46031096, -0.20384297, ..., -0.43297682,
-2.12979052, -2.67217602],
[-2.75695017, -2.14465449, -1.17688786, ..., -0.32599322,
-2.16739012, -2.65226031],
[-2.70533918, -2.49643317, -1.86950641, ..., -0.47782879,
-0.79878241, -2.66414405]],
[[-1.37441681, -0.54965217, -0.0288177 , ..., -0.69535146,
-2.61590546, -2.0004016 ],
[-0.74826047, -0.25727267, -0.46933205, ..., -1.04634275,
-0.99891198, -1.18301087],
[-0.44645302, -0.4857526 , -0.46387903, ..., -0.91002063,
-1.51703446, -0.70378294],
...
[ 0.25470515, 0.30010859, -0.7078344 , ..., 0.95884098,
0.87257852, 0.78445248],
[ 0.89541454, 0.87541371, 0.1233215 , ..., 0.95355904,
0.26139252, 0.7353009 ],
[ 0.42963743, 1.29084385, 1.57845836, ..., 0.58552517,
0.51588286, -0.1126604 ]],
[[-0.87630078, -0.99570863, -0.36829142, ..., 0.205182 ,
-0.74414174, -0.65904228],
[-0.86895441, -0.75981769, -0.09344307, ..., -0.2798458 ,
-0.6485577 , -0.73328435],
[-0.73357821, -0.39963175, 0.27659896, ..., -0.65232246,
-0.82138474, -0.91522523],
...,
[-0.88238532, -0.07674878, 0.66433443, ..., 0.63885238,
-0.14336659, -0.57637156],
[-0.38432406, 0.35059192, 0.55211343, ..., 0.90195504,
-0.39866761, -0.57860287],
[-0.47812988, -0.00373139, 0.40650769, ..., 0.82294193,
0.27791956, -0.71958061]]])
Coordinates:
* lat (lat) float32 -32.5 -27.5 -22.5 -17.5 -12.5 ... 17.5 22.5 27.5 32.5
* lon (lon) float32 2.5 7.5 12.5 17.5 22.5 ... 342.5 347.5 352.5 357.5
* time (time) int64 1850 1851 1852 1853 1854 ... 2014 2015 2016 2017 2018
Attributes:
annualized: 1Field validation#
[15]:
# validate the prior against HadCRUT
stat = 'corr'
valid_fd = job.prior['tas'].compare(
target, stat=stat,
timespan=(1874, 2000),
)
fig, ax = valid_fd.plot(
title=f'{stat}(prior, obs), mean={valid_fd.geo_mean().value[0,0]:.2f}',
projection='PlateCarree',
latlon_range=(-32, 32, 0, 360),
plot_cbar=False,
)
cfr.showfig(fig)
cfr.savefig(fig, f'./figs/pda_{stat}_prior_obs.pdf')
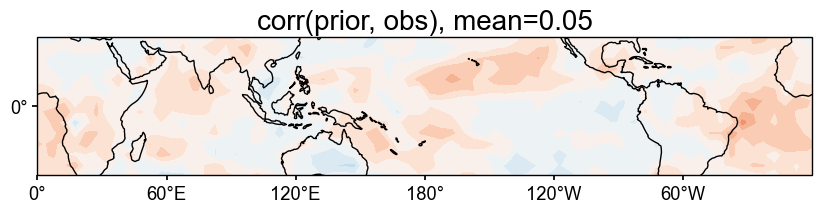
Figure saved at: "figs/pda_corr_prior_obs.pdf"
[16]:
# validate the reconstruction against HadCRUT
valid_fd = res.recons['tas'].compare(
target, stat=stat,
timespan=(1874, 2000),
)
valid_fd.plot_kwargs.update({'cbar_orientation': 'horizontal', 'cbar_pad': 0.1})
fig, ax = valid_fd.plot(
title=f'{stat}(recon, obs), mean={valid_fd.geo_mean().value[0,0]:.2f}',
projection='PlateCarree',
latlon_range=(-32, 32, 0, 360),
plot_cbar=True, plot_proxydb=True, proxydb=job.proxydb.filter(by='tag', keys=['calibrated']),
plot_proxydb_lgd=True, proxydb_lgd_kws={'loc': 'lower left', 'bbox_to_anchor': (1, 0)},
)
cfr.showfig(fig)
cfr.savefig(fig, f'./figs/pda_{stat}_recon_obs.pdf')
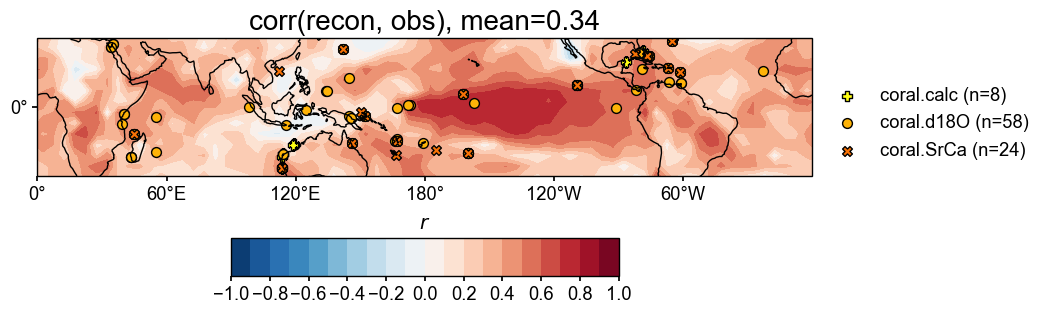
Figure saved at: "figs/pda_corr_recon_obs.pdf"
Timeseries validation#
[17]:
bc09 = cfr.EnsTS().fetch('BC09_NINO34')
bc09_ann = bc09.annualize(months=[12, 1, 2])
[19]:
fig, ax = res.recons['nino3.4'].compare(bc09_ann, timespan=(1874, 2000), ref_name='BC09').plot_qs()
ax.set_xlim(1600, 2000)
ax.set_ylabel('NINO3.4 [K]')
cfr.showfig(fig)
cfr.savefig(fig, f'./figs/pda_corr_recon_BC09.pdf')
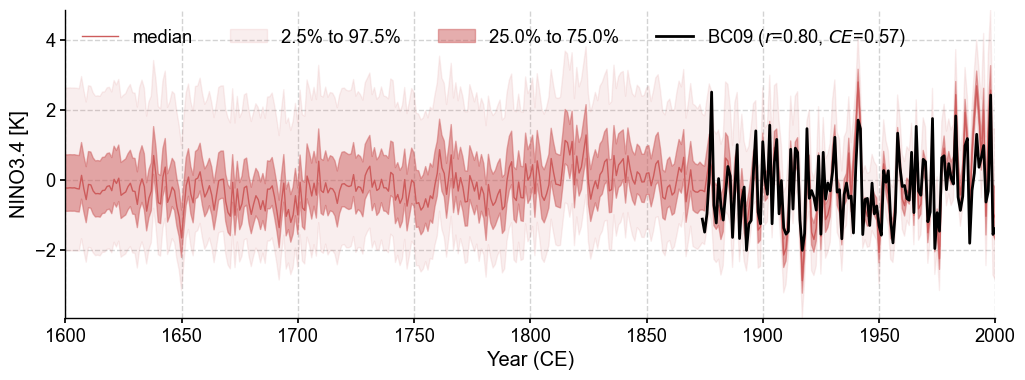
Figure saved at: "figs/pda_corr_recon_BC09.pdf"
[ ]:
